Bronchoalveolar Lavage Fluid-Isolated Biomarkers for the Diagnostic and Prognostic Assessment of Lung Cancer
Abstract
1. Introduction
2. BALF Biomarkers for the Diagnosis and Prognosis of Lung Cancer
2.1. Genetic Biomarkers
2.1.1. EGFR Mutations
2.1.2. KRAS Mutations
2.1.3. ALK Translocations
2.2. Epigenetic Biomarkers
2.3. Post-Transcriptional Biomarkers
2.4. Post-Translational Biomarkers
2.4.1. Proteins
2.4.2. Cell Epitopes
2.5. Metabolite Biomarkers
2.6. Metagenomic Biomarkers
3. BALF Biomarkers for the Identification of Adverse Events of Lung Cancer Treatment
3.1. Pulmonary Infections
3.2. Therapy-Induced Pulmonary Toxicity
4. The Value of Repeated BALF Examination in the Course of Lung Cancer
5. Advantages and Disadvantages of BALF Biomarker Testing in Lung Cancer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Howlader, N.; Noone, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.; et al. SEER Cancer Statistics Review, 1975–2018; National Cancer Institute: Bethesda, MD, USA, 2021.
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef]
- Domagala-Kulawik, J. The relevance of bronchoalveolar lavage fluid analysis for lung cancer patients. Expert Rev. Respir. Med. 2020, 14, 329–337. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, E.Y.; Kim, T.; Chang, Y.S. Compared to plasma, bronchial washing fluid shows higher diagnostic yields for detecting EGFR-TKI sensitizing mutations by ddPCR in lung cancer. Respir. Res. 2020, 21, 142. [Google Scholar] [CrossRef] [PubMed]
- Wongsurakiat, P.; Wongbunnate, S.; Dejsomritrutai, W.; Charoenratanakul, S.; Tscheikuna, J.; Youngchaiyud, P.; Pushpakom, R.; Maranetra, N.; Nana, A.; Chierakul, N.; et al. Diagnostic value of bronchoalveolar lavage and postbronchoscopic sputum cytology in peripheral lung cancer. Respirology 1998, 3, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Binesh, F.; Pirdehghan, A.; Mirjalili, M.R.; Samet, M.; Majomerd, Z.A.; Akhavan, A. Comparative assessment of the diagnostic value of transbronchial lung biopsy and bronchoalveolar lavage fluid cytology in lung cancer. Asian Pac. J. Cancer Prev. 2015, 16, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.S.; Hui, A.B.-Y.; Chabon, J.J.; Esfahani, M.S.; Stehr, H.; Nabet, B.Y.; Zhou, L.; Chaudhuri, A.A.; Benson, J.; Ayers, K.; et al. Genomic profiling of bronchoalveolar lavage fluid in lung cancer. Cancer Res. 2022, 82, 2838. [Google Scholar] [CrossRef]
- Roncarati, R.; Lupini, L.; Miotto, E.; Saccenti, E.; Mascetti, S.; Morandi, L.; Bassi, C.; Rasio, D.; Callegari, E.; Conti, V.; et al. Molecular testing on bronchial washings for the diagnosis and predictive assessment of lung cancer. Mol. Oncol. 2020, 14, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, A.; Fukumitsu, C.; Taira, T.; Abe, H.; Takase, Y.; Murata, K.; Yamaguchi, T.; Azuma, K.; Ishii, H.; Takamori, S.; et al. Epidermal growth factor receptor mutation status in cell-free DNA supernatant of bronchial washings and brushings. Cancer Cytopathol. 2015, 123, 620–628. [Google Scholar] [CrossRef]
- Yanev, N.; Mekov, E.; Valev, D.; Yankov, G.; Milanov, V.; Bichev, S.; Gabrovska, N.; Kostadinov, D. EGFR mutation status yield from bronchoalveolar lavage in patients with primary pulmonary adenocarcinoma compared to a venous blood sample and tissue biopsy. PeerJ 2021, 9, e11448. [Google Scholar] [CrossRef]
- Prim, N.; Quoix, E.; Beau-Faller, M. Tumor cell content for selection of molecular techniques for T790M EGFR mutation detection in non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 1615–1616. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Hur, J.Y.; Lee, K.Y.; Lee, J.C.; Rho, J.K.; Shin, S.H.; Choi, C.-M. Assessment of EGFR mutation status using cell-free DNA from bronchoalveolar lavage fluid. Clin. Chem. Lab. Med. 2017, 55, 1489–1495. [Google Scholar] [CrossRef]
- Kiura, K.; Yoh, K.; Katakami, N.; Nogami, N.; Kasahara, K.; Takahashi, T.; Okamoto, I.; Cantarini, M.; Hodge, R.; Uchida, H. Osimertinib in patients with epidermal growth factor receptor T790M advanced non-small cell lung cancer selected using cytology samples. Cancer Sci. 2018, 109, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Kim, H.J.; Lee, J.S.; Choi, C.-M.; Lee, J.C.; Jung, M.K.; Pack, C.G.; Lee, K.Y. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol. Cancer 2018, 17, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Minna, J.D. Molecular detection of early lung cancer. J. Natl. Cancer Inst. 1999, 91, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Ahrendt, S.A.; Chow, J.T.; Xu, L.-H.; Yang, S.C.; Eisenberger, C.F.; Esteller, M.; Herman, J.G.; Wu, L.; Decker, P.A.; Jen, J.; et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J. Natl. Cancer Inst. 1999, 91, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Oshita, F.; Nomura, I.; Yamada, K.; Kato, Y.; Tanaka, G.; Noda, K. Detection of K-ras mutations of bronchoalveolar lavage fluid cells aids the diagnosis of lung cancer in small pulmonary lesions. Clin. Cancer Res. 1999, 5, 617–620. [Google Scholar] [PubMed]
- Li, J.; Hu, Y.-M.; Wang, Y.; Tang, X.-P.; Shi, W.-L.; Du, Y.-J. Gene mutation analysis in non-small cell lung cancer patients using bronchoalveolar lavage fluid and tumor tissue as diagnostic markers. Int. J. Biol. Markers 2014, 29, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, S.; Seike, M.; Miyanaga, A.; Chiba, M.; Matsuda, K.; Kobayashi, K.; Takahashi, A.; Takeuchi, S.; Minegishi, Y.; Kubota, K.; et al. RT-PCR for Detecting ALK Translocations in Cytology Samples from Lung Cancer Patients. Anticancer Res. 2017, 37, 3295. [Google Scholar] [PubMed]
- Kim, H.; Kwon, Y.M.; Kim, J.S.; Lee, H.; Park, J.-H.; Shim, Y.M.; Han, J.; Park, J.; Kim, D.-H. Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J. Clin. Oncol. 2004, 22, 2363–2370. [Google Scholar] [CrossRef]
- Destro, A.; Bianchi, P.; Alloisio, M.; Laghi, L.; di Gioia, S.; Malesci, A.; Cariboni, U.; Gribaudi, G.; Bulfamante, G.; Marchetti, A.; et al. K-ras and p16INK4A alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer 2004, 44, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Konno, S.; Morishita, Y.; Fukasawa, M.; Shu, Y.; Wang, D.; Tanaka, R.; Minami, Y.; Iijima, T.; Noguchi, M. Anthracotic index and DNA methylation status of sputum contents can be used for identifying the population at risk of lung carcinoma. Cancer Cytopathol. 2004, 102, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, G.; Raji, O.Y.; Markopoulou, S.; Gosney, J.R.; Bryan, J.; Warburton, C.; Walshaw, M.; Sheard, J.; Field, J.K.; Liloglou, T. DNA Methylation Biomarkers Offer Improved Diagnostic Efficiency in Lung Cancer. Cancer Res. 2012, 72, 5692–5701. [Google Scholar] [CrossRef]
- Dietrich, D.; Kneip, C.; Raji, O.; Liloglou, T.; Seegebarth, A.; Schlegel, T.; Flemming, N.; Rausch, S.; Distler, J.; Fleischhacker, M.; et al. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int. J. Oncol. 2012, 40, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yu, W.; Wang, L.; Zhao, M.; Guo, Q.; Lv, S.; Hu, X.; Lou, J. DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J. Cancer 2017, 8, 3585. [Google Scholar] [CrossRef] [PubMed]
- Um, S.-W.; Kim, Y.; Lee, B.B.; Kim, D.; Lee, K.-J.; Kim, H.K.; Han, J.; Kim, H.; Shim, Y.M.; Kim, D.-H. Genome-wide analysis of DNA methylation in bronchial washings. Clin. Epigenetics 2018, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, Z.; Yang, S.; Yang, H.; Jin, J.; Zhu, Y.; Tao, J.; Chen, S.; Xu, J.; Liu, Y.; et al. Diagnosis of pulmonary nodules by DNA methylation analysis in bronchoalveolar lavage fluids. Clin. Epigenetics 2021, 13, 185. [Google Scholar] [CrossRef]
- Tuo, L.; Sha, S.; Huayu, Z.; Du, K. P16INK4a gene promoter methylation as a biomarker for the diagnosis of non-small cell lung cancer: An updated meta-analysis. Thorac. Cancer 2018, 9, 1032–1040. [Google Scholar] [CrossRef]
- Molina-Pinelo, S.; Pastor, M.D.; Suarez, R.; Romero-Romero, B.; González De la Peña, M.; Salinas, A.; García-Carbonero, R.; De Miguel, M.J.; Rodríguez-Panadero, F.; Carnero, A.; et al. MicroRNA clusters: Dysregulation in lung adenocarcinoma and COPD. Eur. Respir. J. 2014, 43, 1740. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, P.; Li, X.-Q.; Bao, Q.-L.; Dai, C.-H.; Ge, L.-P. Elevated levels of survivin and livin mRNA in bronchial aspirates as markers to support the diagnosis of lung cancer. Int. J. Cancer 2013, 132, 1098–1104. [Google Scholar] [CrossRef]
- Rehbein, G.; Schmidt, B.; Fleischhacker, M. Extracellular microRNAs in bronchoalveolar lavage samples from patients with lung diseases as predictors for lung cancer. Clin. Chim. Acta 2015, 450, 78–82. [Google Scholar] [CrossRef]
- Kim, J.E.; Eom, J.S.; Kim, W.-Y.; Jo, E.J.; Mok, J.; Lee, K.; Kim, K.U.; Park, H.-K.; Lee, M.K.; Kim, M.-H. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thorac. Cancer 2018, 9, 911–915. [Google Scholar] [CrossRef]
- Kim, J.O.; Gazala, S.; Razzak, R.; Guo, L.; Ghosh, S.; Roa, W.H.; Bédard, E.L.R. Non-small Cell Lung Cancer Detection Using MicroRNA Expression Profiling of Bronchoalveolar Lavage Fluid and Sputum. Anticancer Res. 2015, 35, 1873. [Google Scholar] [PubMed]
- Li, H.; Jiang, Z.; Leng, Q.; Bai, F.; Wang, J.; Ding, X.; Li, Y.; Zhang, X.; Fang, H.; Yfantis, H.G.; et al. A prediction model for distinguishing lung squamous cell carcinoma from adenocarcinoma. Oncotarget 2017, 8, 50704–50714. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Bovio, E.; Rena, O.; Rrapaj, E.; Mercalli, F.; Veggiani, C.; Paganotti, A.; Andorno, S.; Boldorini, R. Prognostic impact of a 3-MicroRNA signature in cytological samples of small cell lung cancer. Cancer Cytopathol. 2016, 124, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Silva, J.; López-Alfonso, A.; López-Muñiz, M.B.; Peña, C.; Domínguez, G.; García, J.M.; López-Gónzalez, A.; Méndez, M.; Provencio, M.; et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer 2014, 53, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-H.S.; Liu, C.-Y.; Pavlidis, S.; Lo, Y.-L.; Wang, Y.-W.; Chen, C.-H.; Ko, H.-W.; Chung, F.-T.; Lin, T.-Y.; Wang, T.-Y.; et al. Unique Immune Gene Expression Patterns in Bronchoalveolar Lavage and Tumor Adjacent Non-Neoplastic Lung Tissue in Non-Small Cell Lung Cancer. Front. Immunol. 2018, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Macchia, V.; Mariano, A.; Cavalcanti, M.; Coppa, A.; Cecere, C.; Fraioli, G.; Elia, S.; Ferrante, G. Tumor markers and lung cancer: Correlation between serum and bronchial secretion levels of CEA, TPA, CanAg CA-50, NSE and ferritin. Int. J. Biol. Markers 1987, 2, 151–156. [Google Scholar] [CrossRef]
- Edge, S.; Byrd, D.; Compton, C.; Fritz, A.; Greene, F.; Trotti, A. (Eds.) AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Naumnik, W.; Naumnik, B.; Niewiarowska, K.; Ossolinska, M.; Chyczewska, E. Novel cytokines: IL-27, IL-29, IL-31 and IL-33. Can they be useful in clinical practice at the time diagnosis of lung cancer? Exp. Oncol. 2012, 34, 348–353. [Google Scholar]
- Naumnik, W.; Naumnik, B.; Niklińska, W.; Ossolińska, M.; Chyczewska, E. Clinical implications of hepatocyte growth factor, interleukin-20, and interleukin-22 in serum and bronchoalveolar fluid of patients with non-small cell lung cancer. In Advancements in Clinical Research; Pokorski, M., Ed.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2016; pp. 41–49. [Google Scholar]
- Kontakiotis, T.; Katsoulis, K.; Hagizisi, O.; Kougioulis, M.; Gerou, S.; Papakosta, D. Bronchoalveolar lavage fluid alteration in antioxidant and inflammatory status in lung cancer patients. Eur. J. Intern. Med. 2011, 22, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Jakubowska, K.; Naumnik, W.; Niklińska, W.; Chyczewska, E. Clinical significance of HMGB-1 and TGF-β level in serum and BALF of advanced non-small cell lung cancer. In Respiratory Carcinogenesis; Pokorski, M., Ed.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2015; pp. 49–58. [Google Scholar]
- Chen, Z.; Xu, Z.; Sun, S.; Yu, Y.; Lv, D.; Cao, C.; Deng, Z. TGF-β1, IL-6 and TNF-α in Bronchoalveolar Lavage Fluid: Useful Markers for Lung Cancer? Sci. Rep. 2014, 4, 5595. [Google Scholar] [CrossRef]
- Xiong, W.; Ding, W.; Xu, M.; Pudasaini, B.; Sun, J.; Zhao, Y. The screening role of a biomarker panel in BALF among patients with cancer-suspected pulmonary nodules less than 8 mm. Clin. Respir. J. 2020, 14, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Charpidou, A.; Gkiozos, I.; Konstantinou, M.; Eleftheraki, A.; Demertzis, P.; Harrington, K.; Polyzos, A.; Syrigos, K.N. Bronchial washing levels of vascular endothelial growth factor receptor-2 (VEGFR2) correlate with overall survival in NSCLC patients. Cancer Lett. 2011, 304, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Sun, S.F.; Lv, D.; Chen, Z.B.; Ding, Q.L.; Deng, Z.C. Utility of VEGF and sVEGFR-1 in bronchoalveolar lavage fluid for differential diagnosis of primary lung cancer. Asian Pac. J. Cancer Prev. 2013, 14, 2443–2446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, L.; Li, Q.; Zhou, X.-D.; Shi, Y.; Yang, L.; Xu, S.-L.; Chen, C.; Cui, Y.-H.; Zhang, X.; Bian, X.-W. Increased pro-angiogenic factors, infiltrating neutrophils and CD163+ macrophages in bronchoalveolar lavage fluid from lung cancer patients. Int. Immunopharmacol. 2014, 20, 74–80. [Google Scholar] [CrossRef]
- Pio, R.; Garcia, J.; Corrales, L.; Ajona, D.; Fleischhacker, M.; Pajares, M.J.; Cardenal, F.; Seijo, L.; Zulueta, J.J.; Nadal, E.; et al. Complement factor H is elevated in bronchoalveolar lavage fluid and sputum from patients with lung cancer. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2665–2672. [Google Scholar] [CrossRef]
- Ajona, D.; Pajares, M.J.; Corrales, L.; Perez-Gracia, J.L.; Agorreta, J.; Lozano, M.D.; Torre, W.; Massion, P.P.; de-Torres, J.P.; Jantus-Lewintre, E.; et al. Investigation of Complement Activation Product C4d as a Diagnostic and Prognostic Biomarker for Lung Cancer. J. Natl. Cancer Inst. 2013, 105, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Okrój, M.; Pajares, M.J.; Agorreta, J.; Lozano, M.D.; Zulueta, J.J.; Verri, C.; Roz, L.; Sozzi, G.; Pastorino, U.; et al. Complement C4d-specific antibodies for the diagnosis of lung cancer. Oncotarget 2018, 9, 6346–6355. [Google Scholar] [CrossRef]
- Li, Q.K.; Shah, P.; Li, Y.; Aiyetan, P.O.; Chen, J.; Yung, R.; Molena, D.; Gabrielson, E.; Askin, F.; Chan, D.W.; et al. Glycoproteomic analysis of bronchoalveolar lavage (BAL) fluid identifies tumor-associated glycoproteins from lung adenocarcinoma. J. Proteome Res. 2013, 12, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Uribarri, M.; Hormaeche, I.; Zalacain, R.; Lopez-Vivanco, G.; Martinez, A.; Nagore, D.; Ruiz-Argüello, M.B. A new biomarker panel in bronchoalveolar lavage for an improved lung cancer diagnosis. J. Thorac. Oncol. 2014, 9, 1504–1512. [Google Scholar] [CrossRef]
- Ortea, I.; Rodríguez-Ariza, A.; Chicano-Gálvez, E.; Arenas Vacas, M.S.; Jurado Gámez, B. Discovery of potential protein biomarkers of lung adenocarcinoma in bronchoalveolar lavage fluid by SWATH MS data-independent acquisition and targeted data extraction. J. Proteom. 2016, 138, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; McDonald, C.F.; Collins, A.L.; Darby, I.A.; Pouniotis, D.S. Quantitative proteomics of bronchoalveolar lavage fluid in lung adenocarcinoma. Cancer Genom. Proteom. 2015, 12, 39–48. [Google Scholar]
- Carvalho, A.S.; Cuco, C.M.; Lavareda, C.; Miguel, F.; Ventura, M.; Almeida, S.; Pinto, P.; de Abreu, T.T.; Rodrigues, L.V.; Seixas, S.; et al. Bronchoalveolar Lavage Proteomics in Patients with Suspected Lung Cancer. Sci. Rep. 2017, 7, 42190. [Google Scholar] [CrossRef]
- Hmmier, A.; O’Brien, M.E.; Lynch, V.; Clynes, M.; Morgan, R.; Dowling, P. Proteomic analysis of bronchoalveolar lavage fluid (BALF) from lung cancer patients using label-free mass spectrometry. BBA Clin. 2017, 7, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, D.; Shu, J.; Wang, L.; Zhang, F.; Zhang, C.; Yu, H.; Chen, M.; Li, Z.; Guo, X. Protein Glycopatterns in Bronchoalveolar Lavage Fluid as Novel Potential Biomarkers for Diagnosis of Lung Cancer. Front. Oncol. 2021, 10, 568433. [Google Scholar] [CrossRef]
- Domagala-Kulawik, J.; Osinska, I.; Hoser, G.J. Mechanisms of immune response regulation in lung cancer. Transl. Lung Cancer 2014, 3, 15. [Google Scholar]
- Kwiecien, I.; Stelmaszczyk-Emmel, A.; Polubiec-Kownacka, M.; Dziedzic, D.; Domagala-Kulawik, J. Elevated regulatory T cells, surface and intracellular CTLA-4 expression and interleukin-17 in the lung cancer microenvironment in humans. Cancer Immunol. Immunother. 2017, 66, 161–170. [Google Scholar] [CrossRef]
- Zhang, B.; Yao, G.; Zhang, Y.; Gao, J.; Yang, B.; Rao, Z.; Gao, J. M2-polarized tumor-associated macrophages are associated with poor prognoses resulting from accelerated lymphangiogenesis in lung adenocarcinoma. Clinics 2011, 66, 1879–1886. [Google Scholar] [CrossRef]
- Osińska, I.; Stelmaszczyk-Emmel, A.; Polubiec-Kownacka, M.; Dziedzic, D.; Domagała-Kulawik, J. CD4+/CD25high/FoxP3+/CD127−regulatory T cells in bronchoalveolar lavage fluid of lung cancer patients. Hum. Immunol. 2016, 77, 912–915. [Google Scholar] [CrossRef]
- Hu, X.; Gu, Y.; Li, D.; Zhao, S.; Hua, S.; Jiang, Y. Analyzing the percentage of different PD-1+ T cell subsets in peripheral blood and bronchoalveolar lavage fluid of small cell lung cancer patients: A prospective study. Clin. Exp. Pharmacol. Physiol. 2019, 46, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gu, Y.; Zhao, S.; Hua, S.; Jiang, Y. Immunotherapy. Increased IL-10+ CD206+ CD14+ M2-like macrophages in alveolar lavage fluid of patients with small cell lung cancer. Cancer Immunol. 2020, 69, 2547–2560. [Google Scholar] [CrossRef] [PubMed]
- Masuhiro, K.; Tamiya, M.; Fujimoto, K.; Koyama, S.; Naito, Y.; Osa, A.; Hirai, T.; Suzuki, H.; Okamoto, N.; Shiroyama, T.; et al. Bronchoalveolar lavage fluid reveals factors contributing to the efficacy of PD-1 blockade in lung cancer. JCI Insight 2022, 7, e157915. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.-M.; Ward, T.H.; Blackhall, F.H.; Dive, C. Circulating tumour cells: Their utility in cancer management and predicting outcomes. Ther. Adv. Med. Oncol. 2010, 2, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ge, F.; Cui, W.; Wang, F.; Yang, Z.; Guo, Y.; Li, L.; Bremner, R.M.; Lin, P.P. Lung cancer circulating tumor cells isolated by the EpCAM-independent enrichment strategy correlate with Cytokeratin 19-derived CYFRA21-1 and pathological staging. Clin. Chim. Acta 2013, 419, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-P.; Liu, S.-H.; Chen, C.-T.; Lv, L.; Li, D.; Liu, Q.-Y.; Liu, G.-L.; Wu, Y. Circulating tumor cells as a new predictive and prognostic factor in patients with small cell lung cancer. J. Cancer 2020, 11, 2113. [Google Scholar] [CrossRef]
- Yang, D.; Yang, X.; Li, Y.; Zhao, P.; Fu, R.; Ren, T.; Hu, P.; Wu, Y.; Yang, H.; Guo, N. Clinical significance of circulating tumor cells and metabolic signatures in lung cancer after surgical removal. J. Transl. Med. 2020, 18, 1–13. [Google Scholar] [CrossRef]
- Zhong, M.; Zhang, Y.; Pan, Z.; Wang, W.; Zhang, Y.; Weng, Y.; Huang, H.; He, Y.; Liu, O. Clinical Utility of Circulating Tumor Cells in the Early Detection of Lung Cancer in Patients with a Solitary Pulmonary Nodule. Technol. Cancer Res. Treat. 2021, 20, 15330338211041465. [Google Scholar] [CrossRef]
- Ciebiada, M.; Górski, P.; Antczak, A. Eicosanoids in Exhaled Breath Condensate and Bronchoalveolar Lavage Fluid of Patients with Primary Lung Cancer. Dis. Markers 2012, 32, 562862. [Google Scholar] [CrossRef][Green Version]
- Schmid, S.; Kübler, M.; Korcan Ayata, C.; Lazar, Z.; Haager, B.; Hoßfeld, M.; Meyer, A.; Cicko, S.; Elze, M.; Wiesemann, S.; et al. Altered purinergic signaling in the tumor associated immunologic microenvironment in metastasized non-small-cell lung cancer. Lung Cancer 2015, 90, 516–521. [Google Scholar] [CrossRef]
- Callejón-Leblic, B.; García-Barrera, T.; Grávalos-Guzmán, J.; Pereira-Vega, A.; Gómez-Ariza, J.L. Metabolic profiling of potential lung cancer biomarkers using bronchoalveolar lavage fluid and the integrated direct infusion/ gas chromatography mass spectrometry platform. J. Proteom. 2016, 145, 197–206. [Google Scholar] [CrossRef]
- Lui, G.Y.; Kovacevic, Z.; Richardson, V.; Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget 2015, 6, 18748. [Google Scholar] [CrossRef]
- Callejon-Leblic, B.; Gómez-Ariza, J.L.; Pereira-Vega, A.; Garcia-Barrera, T. Metal dyshomeostasis based biomarkers of lung cancer using human biofluids. Metallomics 2018, 10, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Huang, Y.; Zhang, Z.; Liao, J.; Ding, Y.; Fang, X.; Liu, L.; Luo, J.; Kong, J. A preliminary study of microbiota diversity in saliva and bronchoalveolar lavage fluid from patients with primary bronchogenic carcinoma. Med. Sci. Monit. 2019, 25, 2819. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Z.; Wang, J.; Ding, C.; Sun, C.; Liu, P.; Xu, X.; Liu, Y.; Chen, B.; Gu, B. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 2020, 9, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Sung, J.Y.; Yong, D.; Chun, J.; Kim, S.Y.; Song, J.H.; Chung, K.S.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef]
- Patnaik, S.K.; Cortes, E.G.; Kannisto, E.D.; Punnanitinont, A.; Dhillon, S.S.; Liu, S.; Yendamuri, S. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non–small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2021, 161, 419–429.e16. [Google Scholar] [CrossRef]
- Zheng, L.; Sun, R.; Zhu, Y.; Li, Z.; She, X.; Jian, X.; Yu, F.; Deng, X.; Sai, B.; Wang, L.; et al. Lung microbiome alterations in NSCLC patients. Sci. Rep. 2021, 11, 11736. [Google Scholar] [CrossRef]
- Jang, H.J.; Choi, J.Y.; Kim, K.; Yong, S.H.; Kim, Y.W.; Kim, S.Y.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; Park, M.S.; et al. Relationship of the lung microbiome with PD-L1 expression and immunotherapy response in lung cancer. Respir. Res. 2021, 22, 322. [Google Scholar] [CrossRef]
- Espuela-Ortiz, A.; Lorenzo-Diaz, F.; Baez-Ortega, A.; Eng, C.; Hernandez-Pacheco, N.; Oh, S.S.; Lenoir, M.; Burchard, E.G.; Flores, C.; Pino-Yanes, M. Bacterial salivary microbiome associates with asthma among African American children and young adults. Pediatric Pulmonol. 2019, 54, 1948–1956. [Google Scholar] [CrossRef]
- Molyneaux, P.L.; Cox, M.J.; Wells, A.U.; Kim, H.C.; Ji, W.; Cookson, W.O.; Moffatt, M.F.; Kim, D.S.; Maher, T.M. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir. Res. 2017, 18, 1–6. [Google Scholar] [CrossRef]
- Yu, D.P.; Cheng, X.; Liu, Z.D.; Xu, S.F. Comparative beneficiary effects of immunotherapy against chemotherapy in patients with advanced NSCLC: Meta-analysis and systematic review. Oncol. Lett. 2017, 14, 1568–1580. [Google Scholar] [CrossRef]
- Weber, J.S.; Kähler, K.C.; Hauschild, A. Management of immune-related adverse events and kinetics of response with ipilimumab. J. Clin. Oncol. 2012, 30, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Kyi, C.; Hellmann, M.D.; Wolchok, J.D.; Chapman, P.B.; Postow, M.A. Opportunistic infections in patients treated with immunotherapy for cancer. J. Immunother. Cancer 2014, 2, 19. [Google Scholar] [CrossRef]
- Fujita, K.; Terashima, T.; Mio, T. Anti-PD1 antibody treatment and the development of acute pulmonary tuberculosis. J. Thorac. Oncol. 2016, 11, 2238–2240. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Sandur, S.; Meli, Y.; Arroliga, A.C.; Stoller, J.K.; Mehta, A.C. Role of flexible bronchoscopy in immunocompromised patients with lung infiltrates. Chest 2004, 125, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Rizvi, N.A.; Goldman, J.W.; Gettinger, S.N.; Borghaei, H.; Brahmer, J.R.; Ready, N.E.; Gerber, D.E.; Chow, L.Q.; Juergens, R.A.; et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017, 18, 31–41. [Google Scholar] [CrossRef]
- Suresh, K.; Naidoo, J.; Lin, C.T.; Danoff, S. Immune checkpoint immunotherapy for non-small cell lung cancer: Benefits and pulmonary toxicities. Chest 2018, 154, 1416–1423. [Google Scholar] [CrossRef]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Gettinger, S.N.; Horn, L.; Gandhi, L.; Spigel, D.R.; Antonia, S.J.; Rizvi, N.A.; Powderly, J.D.; Heist, R.S.; Carvajal, R.D.; Jackman, D.M.; et al. Overall survival and long-term safety of nivolumab (anti–programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non–small-cell lung cancer. J. Clin. Oncol. 2015, 33, 2004. [Google Scholar] [CrossRef]
- Delaunay, M.; Cadranel, J.; Lusque, A.; Meyer, N.; Gounant, V.; Moro-Sibilot, D.; Michot, J.-M.; Raimbourg, J.; Girard, N.; Guisier, F.; et al. Immune-checkpoint inhibitors associated with interstitial lung disease in cancer patients. Eur. Respir. J. 2017, 50, 1700050. [Google Scholar] [CrossRef]
- Suresh, K.; Naidoo, J.; Zhong, Q.; Xiong, Y.; Mammen, J.; de Flores, M.V.; Cappelli, L.; Balaji, A.; Palmer, T.; Forde, P.M.; et al. The alveolar immune cell landscape is dysregulated in checkpoint inhibitor pneumonitis. J. Clin. Investig. 2019, 129, 4305–4315. [Google Scholar] [CrossRef]
- Chopra, A.; Nautiyal, A.; Kalkanis, A.; Judson, M.A. Drug-induced sarcoidosis-like reactions. Chest 2018, 154, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Gkiozos, I.; Kopitopoulou, A.; Kalkanis, A.; Vamvakaris, I.N.; Judson, M.A.; Syrigos, K.N. Sarcoidosis-like reactions induced by checkpoint inhibitors. J. Thorac. Oncol. 2018, 13, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- Costabel, U.; Hunninghake, G. ATS/ERS/WASOG statement on sarcoidosis. Sarcoidosis statement committee. American thoracic society. European respiratory society. World association for sarcoidosis and other granulomatous disorders. Eur. Respir. J. 1999, 14, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Montaudié, H.; Pradelli, J.; Passeron, T.; Lacour, J.P.; Leroy, S. Pulmonary sarcoid-like granulomatosis induced by nivolumab. Br. J. Dermatol. 2017, 176, 1060–1063. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, L.; Ji, W.; Wang, X.; Zhu, X.; Hayman, J.A.; Kalemkerian, G.P.; Yang, W.; Brenner, D.; Lawrence, T.S.; et al. Elevation of plasma TGF-β1 during radiation therapy predicts radiation-induced lung toxicity in patients with non-small-cell lung cancer: A combined analysis from Beijing and Michigan. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1385–1390. [Google Scholar] [CrossRef]
- Chen, Y.; Rubin, P.; Williams, J.; Hernady, E.; Smudzin, T.; Okunieff, P. Circulating IL-6 as a predictor of radiation pneumonitis. Int. J. Radiat. Oncol. Biol. Phys. 2001, 49, 641–648. [Google Scholar] [CrossRef]
- Park, K.-J.; Oh, Y.-T.; Kil, W.-J.; Park, W.; Kang, S.-H.; Chun, M. Bronchoalveolar lavage findings of radiation induced lung damage in rats. J. Radiat. Res. 2009, 50, 177–182. [Google Scholar] [CrossRef][Green Version]
- Hong, J.-H.; Jung, S.-M.; Tsao, T.C.Y.; Wu, C.-J.; Lee, C.-Y.; Chen, F.-H.; Hsu, C.-H.; Mcbride, W.H.; Chiang, C.-S. Bronchoalveolar lavage and interstitial cells have different roles in radiation-induced lung injury. Int. J. Radiat. Biol. 2003, 79, 159–167. [Google Scholar] [CrossRef]
- Toma, C.L.; Serbescu, A.; Alexe, M.; Cervis, L.; Ionita, D.; Bogdan, M.A. The bronchoalveolar lavage pattern in radiation pneumonitis secondary to radiotherapy for breast cancer. Maedica 2010, 5, 250. [Google Scholar]
- Crohns, M.; Saarelainen, S.; Laine, S.; Poussa, T.; Alho, H.; Kellokumpu-Lehtinen, P. Cytokines in bronchoalveolar lavage fluid and serum of lung cancer patients during radiotherapy—Association of interleukin-8 and VEGF with survival. Cytokine 2010, 50, 30–36. [Google Scholar] [CrossRef]
- Yamagishi, T.; Kodaka, N.; Kurose, Y.; Watanabe, K.; Nakano, C.; Kishimoto, K.; Oshio, T.; Niitsuma, K.; Matsuse, H. Analysis of predictive parameters for the development of radiation-induced pneumonitis. Ann. Thorac. Med. 2017, 12, 252. [Google Scholar] [PubMed]
- Aso, S.; Navarro-Martin, A.; Castillo, R.; Padrones, S.; Castillo, E.; Montes, A.; Martínez, J.I.; Cubero, N.; López, R.; Rodríguez, L.; et al. Severity of radiation pneumonitis, from clinical, dosimetric and biological features: A pilot study. Radiat. Oncol. 2020, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zarogoulidis, P.; Rapti, A.; Sardeli, C.; Chinelis, P.; Athanasiadou, A.; Paraskevaidou, K.; Kallianos, A.; Veletza, L.; Trakada, G.; Hohenforst-Schmidt, W.; et al. Re-biopsy after relapse of targeted therapy. T790M after epidermal growth factor mutation, where and why based on a case series. Respir. Med. Case Rep. 2017, 21, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Kenmotsu, H.; Taira, T.; Omori, S.; Nakashima, K.; Wakuda, K.; Ono, A.; Naito, T.; Murakami, H.; Mori, K.; et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci. 2016, 107, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
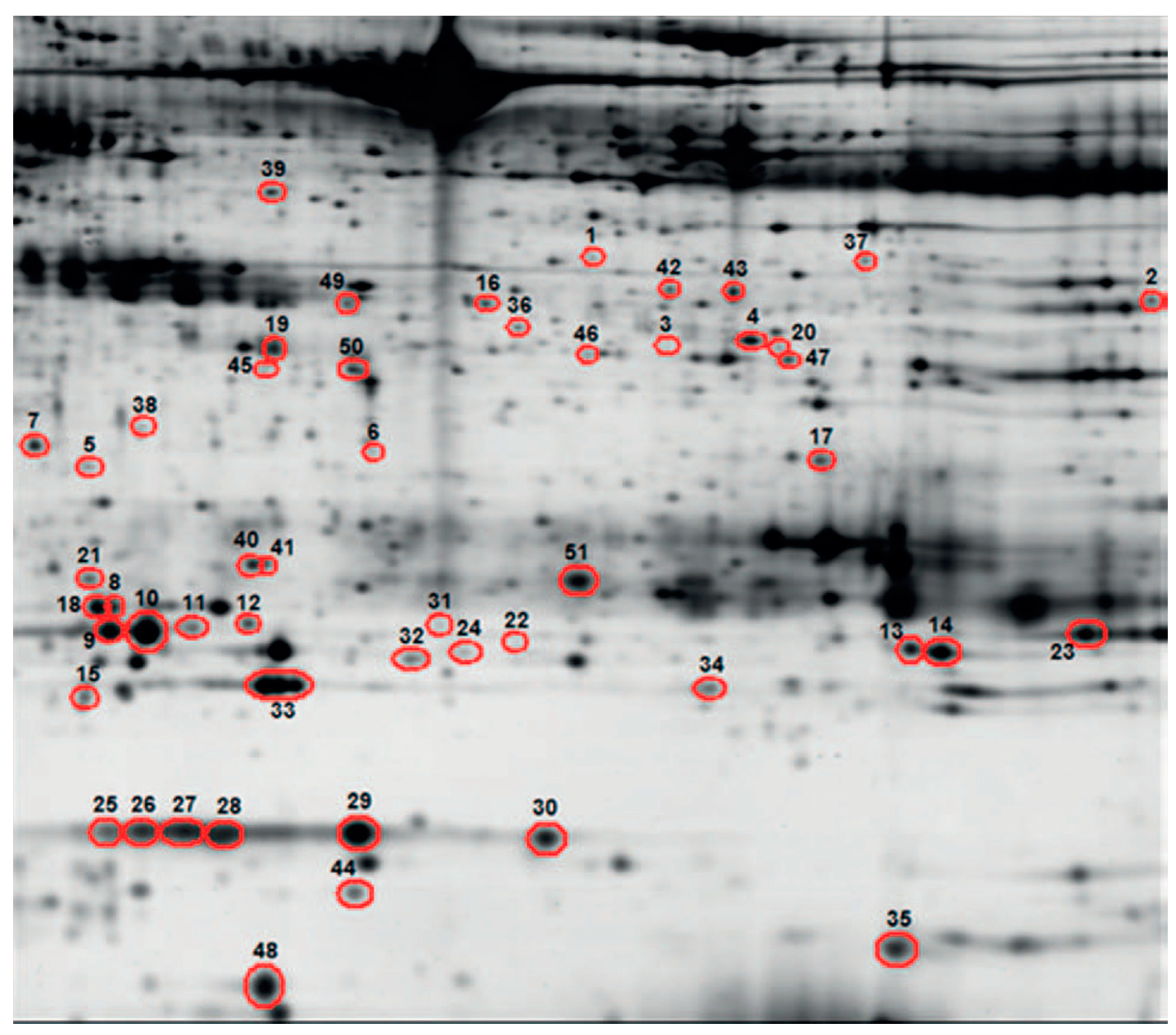
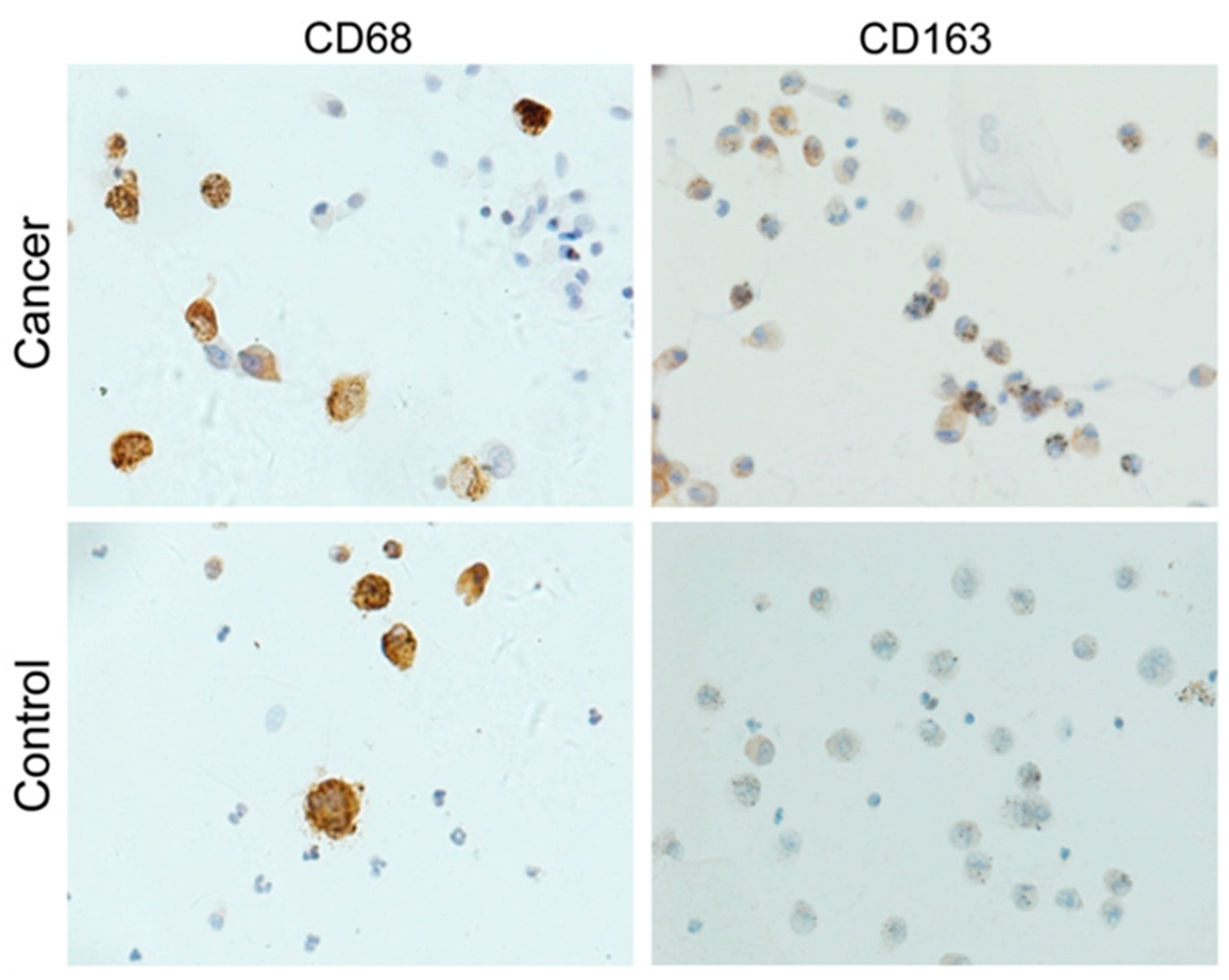
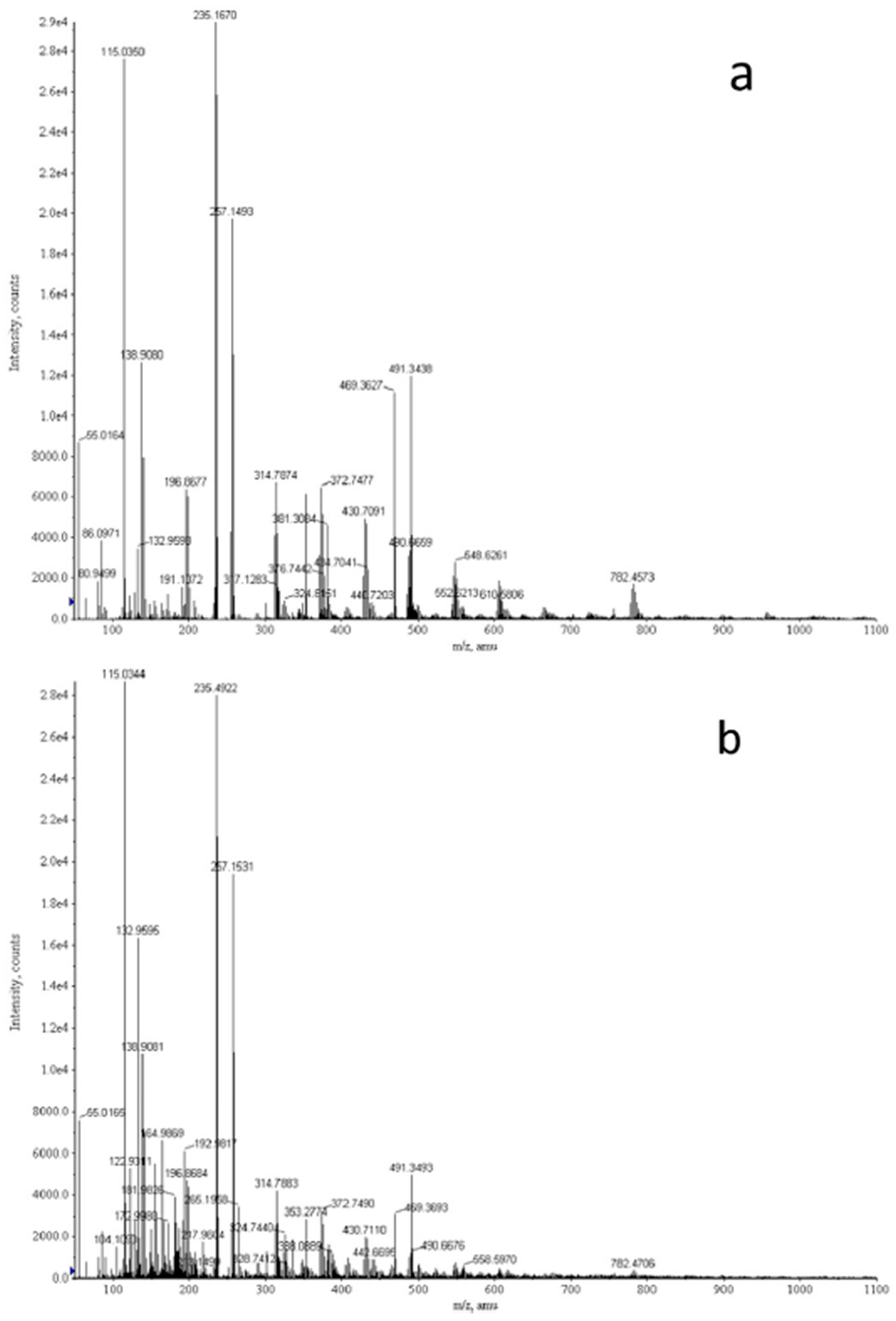
| Study | Patients | Controls | Processing | Detection | Biomarker | Role | Accuracy |
|---|---|---|---|---|---|---|---|
| Genetic/epigenetic biomarkers | |||||||
| Lee 2020 [4] | n = 73 (M 38) Mean age: 65.3 ± 9.8 y Type: AC 65, SQCC 7, other 1 Stage: I 20, II 10, III 10, IV 27 | - | Cell-free DNA from BALF supernatant | Droplet digital allele-specific PCR | EGFR L858R mutation | Prediction of tumor molecular profile | AUC 0.96 Acc 95% |
| EGFR E19del mutation | AUC 0.86 Acc 85% | ||||||
| Nair 2022 [7] | n = 35 (M 18) Age: 40–83 y Type: AC 30, SQCC 2, NSCLC NOS 1, SCC 1, other 1 Stage: I 18, II 6, III 6, IV 5 | n = 21 (M 12) Age: 46–76 y | Cell-free DNA from BALF supernatant | Deep sequencing | 11 gene feature classifier | LC diagnosis | AUC 0.84 Sens 69% Spec 100% |
| n = 31 | - | Any tumor variant detected | Prediction of tumor molecular profile | Acc 81% | |||
| Roncarati 2020 [8] | n = 91 (M 60) Age: 47–85 y Type: AC 41, SQCC 31, SCC 11, undefined 7 Stage: I 13, II 7, III 25, IV 43 | n = 31 (M 21) Age: 42–86 y | Cellular DNA/RNA from BALF cell pellet | Droplet digital methylation-specific PCR | CDH1 methylation | LC diagnosis | Sens 64% Spec 74% |
| DLC1 methylation | Sens 37% Spec 94% | ||||||
| PRPH methylation | Sens 40% Spec 100% | ||||||
| RASSF1A methylation | Sens 46% Spec 100% | ||||||
| 4-gene methylation panel | AUC 0.93 Sens 97% Spec 74% | ||||||
| - | Next-generation sequencing | ALK fusions | Prediction of tumor molecular profile | Acc 96% | |||
| BRAF V600E mutation | Acc 100% | ||||||
| EGFR mutations | Acc 97% | ||||||
| ERBB2/HER2 mutations | Acc 100% | ||||||
| KRAS mutations | Acc 90% | ||||||
| MET mutations | Acc 100% | ||||||
| ROS1 fusions | Acc 100% | ||||||
| Kawahara 2015 [9] | n = 42 (M 23) Age: 42–84 y Type: AC | - | Cell-free DNA from BALF supernatant | Allele-specific PCR/ FRET-PHFA | EGFR mutations | Prediction of tumor molecular profile | Acc 47% |
| Yanev 2021 [10] | n = 26 (M 13) Mean age: 63.3 y Type: AC | - | Cell-free DNA from BALF supernatant | Allele-specific PCR | EGFR mutations | Prediction of tumor molecular profile | Acc 92% |
| Park 2017 [12] | n = 20 (M 5) Age: 43–77 y Type: AC Stage: I 1, II 1, III 1, IV 17 | - | Cell-free DNA from BALF supernatant | PNA-mediated clamping PCR/ PNA clamping-assisted fluorescence melting curve analysis | EGFR mutations | Prediction of tumor molecular profile | Acc 92% |
| Prediction of treatment response (EGFR TKI) | - | ||||||
| Hur 2018 [14] | n = 23 | - | EV DNA from BALF EV pellet | PNA-mediated clamping PCR | EGFR mutations | Prediction of tumor molecular profile | Acc 100% |
| Cell-free DNA from BALF supernatant | Acc 71% | ||||||
| Ahrendt 1999 [16] | n = 50 Type: AC 25, SQCC 23, other 2 Stage: I 28, II 15, III 7 | - | Cellular DNA from BALF cell pellet | Oligonucleotide plaque hybridization | p53 mutations | Prediction of tumor molecular profile | Acc 39% |
| Allele-specific PCR | KRAS mutations | Acc 50% | |||||
| Methylation-specific PCR | p16 methylation | Acc 63% | |||||
| Microsatellite fragment analysis | 15 microsatellite markers | Acc 14% | |||||
| Combined panel | Acc 53% | ||||||
| Oshita 1999 [17] | n = 20 Type: AC | n = 13 | Cellular DNA from BALF cell pellet | Allele-specific PCR | KRAS codon 12 mutation | LC diagnosis | Sens 79% Spec 64% |
| Li 2014 [18] | n = 48 (M 27) Age: 42–75 y Type: AC 32, SQCC 14, LCC 2 Stage: I 19, II 25, III 4 | n = 26 (M 17) Age: 28–70 y | Cellular DNA from BALF cell pellet | PCR single-strand conformation polymorphism | KRAS mutations | LC diagnosis | Sens 38% Spec 92% |
| Prediction of tumor molecular profile | Acc 72% | ||||||
| p53 mutations | LC diagnosis | Sens 44% Spec 96% | |||||
| Prediction of tumor molecular profile | Acc 75% | ||||||
| Combined panel | LC diagnosis | Sens 67% Spec 89% | |||||
| Prediction of tumor molecular profile | Acc 70% | ||||||
| Nakamichi 2017 [19] | n = 36 | - | Cellular RNA from BALF cell pellet | mRNA-specific reverse transcription-PCR | ALK translocations | Prediction of tumor molecular profile | Acc 97% |
| Kim 2004 [20] | n = 85 (M 57) Mean age: 65 ± 17 y Type: AC 31, SQCC 43, other 11 Stage: I 52, II 33 | n = 127 (M 84) Mean age: 62 ± 14 y | Cellular DNA from BALF cell pellet | Methylation-specific PCR | p16 methylation | LC diagnosis | Sens 16% Spec 94% |
| RARβ methylation | Sens 15% Spec 87% | ||||||
| H-cadherin methylation | Sens 13% Spec 97% | ||||||
| RASSF1A methylation | Sens 18% Spec 96% | ||||||
| Nikolaidis 2012 [23] | n = 139 (M 80) Mean age: 68.4 ± 8.1 y Type: AC 22, SQCC 31, SCC 39, LCC 16, other 20, unknown 11 | n = 109 (M 63) Mean age: 67.6 ± 8.8 y | Cellular DNA from BALF cell pellet | Methylation-specific PCR | Methylation panel (p16, RASSF1, WT1, TERT) | LC diagnosis | Sens 82% Spec 91% |
| Dietrich 2012 [24] | n = 125 (M 72) Age: 46–85 y Type: AC 26, SQCC 28, NSCLC NOS 9, SCC 40, other 22 | n = 125 (M 61) Age: 45–86 y | Cellular DNA from BALF cell pellet | Methylation-specific PCR | SHOX2 methylation | LC diagnosis | AUC 0.94 Sens 78% Spec 96% |
| Zhang 2017 [25] | n = 284 (M 212) Age: 31–85 Type: AC 92, SQCC 107, SCC 42, LCC 5, unknown 38 Stage: I 28, II 30, III 133, IV 93 | n = 38 (M 28) Age: 29–75 y | Cellular DNA from BALF cell pellet | Methylation-specific PCR | Methylation panel (SHOX2, RASSF1A) | LC diagnosis | AUC 0.89 Sens 81% Spec 97% |
| Um 2018 [26] | n = 31 | n = 10 | Cellular DNA from BALF cell pellet | DNA methylation microarray | Methylation panel (TFAP2A, TBX15, PHF11, TOX2, PRR15, PDGFRA, HOXA11) | LC diagnosis | AUC 0.87 Sens 87% Spec 83% |
| Li 2021 [27] | n = 52 (M 33) Type: AC 37, SQCC 10, other 5 Stage: I 34, II 1, III 1, IV 4, unknown 12 | n = 59 (M 33) | Cellular DNA from BALF cell pellet | Methylation-specific PCR | Methylation panel (LHX9, GHSR, HOXA11, PTGER4-2, HOXB4-3) | LC diagnosis | AUC 0.82 Sens 70% Spec 82% |
| Post-transcriptional biomarkers | |||||||
| Molina-Pinelo 2014 [29] | n = 48 (M 40) Type: AC Stage: I-II 2, III 15, IV 31 | n = 16 (M 15) | Cellular RNA from BALF cell pellet | MicroRNA microfluidic card array | Four upregulated miRNA clusters (chromosome loci 13q31.3, 7q22.1, Xq26.2, 11q13.1) | LC diagnosis | - |
| Li 2013 [30] | n = 70 (M 52) Mean age: 64 ± 24 y Type: AC 37, SQCC 25, LCC 4, SCC 4 Stage: I 10, II 24, III 25, IV 11 | n = 26 (M 19) Mean age: 55 ± 19 y | Cellular RNA from BALF cell pellet | mRNA-specific reverse transcription-PCR | Survivin expression ratio > 0.35 | LC diagnosis | AUC 0.83 Sens 83% Spec 96% |
| Livin expression ratio > 0.3 | AUC 0.68 Sens 63% Spec 92% | ||||||
| Rehbein 2015 [31] | n = 30 (M 21) Median age 64.5 y | n = 30 (M 17) Median age 63.5 y | Cell-free RNA from BALF supernatant | MicroRNA microfluidic card array | Five upregulated miRNAs (U6 snRNA, hsa-miR 1285, hsa-miR 1303, hsa-miR 29a-5p, hsa-miR 650) | LC diagnosis | - |
| Kim 2018 [32] | n = 13 (M 7) Age: 47–72 y Type: AC Stage: I 10, II 3 | n = 15 | EV RNA from BALF EV pellet | miRNA-specific reverse transcription-PCR | miR-126 and Let-7a were significantly upregulated | LC diagnosis | - |
| Kim 2015 [33] | n = 21 (M 17) Age: 46–84 y Type: AC 13, SQCC 5, LCC 3 Stage: I 12, II 9 | n = 10 (M 8) Age: 30–77 y | Cellular RNA from BALF cell pellet | miRNA-specific reverse transcription-PCR | High expression cluster of a 5-miRNA panel (miR-21, miR-143, miR-155, miR-210, miR-372) | LC diagnosis | Sens 86% Spec 100% |
| Li 2017 [34] | n = 127 (M 82) Age: 66 ± 8 y Type: AC 45, SQCC 82 Stage: I 52, II 38, III-IV 37 | - | Cellular RNA from BALF cell pellet | Droplet digital miRNA-specific PCR | 2-miRNA (miR-205-5p, miR-944) prediction model | Discrimination of SQCC from AC | AUC 0.997 Sens 95% Spec 97% |
| Mancuso 2016 [35] | n = 50 (M 32) Age: 34–82 y Type: SCC Stage: III 18, IV 32 | - | Cellular RNA from BALF cell pellet | miRNA-specific reverse transcription-PCR | Above median expression levels of a 3-miRNA panel (miR-192, miR-200c, miR-205) | Overall survival (worse) | - |
| Rodríguez 2014 [36] | n = 30 (M 23) Age: 45–83 y Type: AC 14, SQCC 16 | n = 75 (M 46) Age: 18–87 y | EV RNA from BALF EV pellet | MicroRNA real-time PCR array | 10 miRNAs were upregulated and 10 downregulated | LC diagnosis | - |
| Kuo 2018 [37] | n = 34 (M 19) Mean age: 58.5 ± 12.8 y Type: AC 26, SQCC 8 Stage: III 11, IV 23 | n = 14 (M 7) Mean age: 53.3 ± 11.4 y | Cellular RNA from BALF cell pellet | mRNA-specific reverse transcription-PCR | 9-gene (SPP1, CEACAM6, MMP7, SLC40A1, IGJ, IGKC, CPA3, YES1, CXCL13) prediction model | LC diagnosis | AUC 0.92 |
| Post-translational biomarkers (proteins, cell epitopes, metabolites) | |||||||
| Macchia 1987 [38] | n = 37 Type: AC 4, SQCC 23, LCC 3, SCC 7 | n = 20 | Cell-free BALF supernatant | RIA | CEA | LC diagnosis | Sens 57% Spec 65% |
| TPA | Sens 65% Spec 20% | ||||||
| NSE | SCC diagnosis | Sens 71% Spec 90% | |||||
| Ferritin | Sens 71% Spec 100% | ||||||
| CanAg CA-50 | Sens 100% Spec 55% | ||||||
| Naumnik 2012 [40] | n = 45 (M 38) Mean age: 61.9 ± 4 y Type: AC 9, SQCC 22, NSCLC NOS 14 Stage: III 18, IV 27 | n = 15 (M 13) Mean age: 60.1 ± 5 y | Cell-free BALF supernatant | ELISA | IL-27 (↑) | LC diagnosis | - |
| IL-27, IL-29 (↓) | Discrimination of advanced stage | - | |||||
| Naumnik 2016 [41] | n = 46 (M 46) Mean age: 63 ± 3 y Type: AC 10, SQCC 25, LCC 11 Stage: III 20, IV 26 | n = 15 (M 12) Mean age 60 ± 4 y | Cell-free BALF supernatant | ELISA | HGF, IL-22 (↓) | LC diagnosis | - |
| IL-22 (↑) | Overall survival (worse) | - | |||||
| Kontakiotis 2011 [42] | n = 42 (M 42) Age: 43–80 y Type: AC 7, SQCC 22, SCC 10, other 3 | n = 16 (M 16) Age: 45–77 y | Cell-free BALF supernatant | ELISA | TNF-α (↑) | LC diagnosis | - |
| Colorimetric assay | Total antioxidants, glutathione (↑) | - | |||||
| Jakubowska 2015 [43] | n = 45 (M 38) Mean age: 61.7 ± 8.3 y Type: AC 20, SQCC 22, LCC 3 Stage: III 18, IV 27 | n = 15 (M 13) Mean age: 60.1 ± 5.0 y | Cell-free BALF supernatant | ELISA | TGF-β (↑) | LC diagnosis | - |
| Chen 2014 [44] | n = 45 (M 28) Mean age: 60.8 ± 1.2 y Type: AC 11, SQCC 18, SCC 10, other 6 | n = 33 (M 19) Mean age: 58.2 ± 1.7 y | Cell-free BALF supernatant | ELISA | TGF-β1 >10.85 pg/ml | LC diagnosis | AUC 0.7 Sens 62% Spec 61% |
| Xiong 2020 [45] | n = 219 (M 150) Mean age: 68.4 ± 18.8 y Type: AC 136, SQCC 43, SCC 35, other 5 Stage: 0 38, I 93, II 50, III 28, IV 10 | n = 186 (M 125) Mean age: 40.6 ± 15.5 y | Cell-free BALF supernatant | ELISA | VEGF >234.1 pg/mL, TGF-β >81.8 pg/mL, HGF 44.6 pg/mL (at least 2 positive) | LC diagnosis | AUC 0.81 Sens 82% Spec 61% |
| Charpidou 2011 [46] | n = 40 (M 37) Age: 45–82 y Type: AC 12, SQCC 19, other 9 Stage: I 3, III 14, IV 23 | - | Cell-free BALF supernatant | ELISA | VEGF (↓) | Prediction of treatment response (chemotherapy) | - |
| VEGFR1 >53.2 pg/ml | Progression free survival (worse) | - | |||||
| VEGFR2 >705.3 pg/ml | Overall survival (worse) | - | |||||
| Cao 2013 [47] | n = 37 (M 28) Mean age: 55.4 ± 8.4 y Type: AC 15, SQCC 19, SCC 3 Stage: I 23, II 9, III 5 | n = 19 (M 12) Mean age: 48.1 ± 9.2 y | Cell-free BALF supernatant | ELISA | VEGF >214 pg/ml | LC diagnosis | AUC 0.86 Sens 82% Spec 84% |
| Chen 2014 [48] | n = 54 (M 60) Median age: 60 y Type: AC 9, SQCC 36, SCC 9 | n = 12 (M 6) Median age: 37 y | Cell-free BALF supernatant | ELISA | IL-8, VEGF (↑) | LC diagnosis | - |
| Pio 2010 [49] | n = 56 (M 45) Age: 38–83 y Type: AC 12, SQCC 24, LCC 4, SCC 9, other 7 | n = 22 (M 14) Age: 30–82 y | Cell-free BALF supernatant | ELISA | Complement factor H >1 μg/mL | LC diagnosis | Sens 62% Spec 77% |
| Albumin >17 μg/mL | Sens 68% Spec 71% | ||||||
| Ajona 2013 [50] | n = 50 (M 41) Type: AC 12, SQCC 22, SCC 9, other 7 | n = 22 (M 14) | Cell-free BALF supernatant | ELISA | Complement C4-derived fragments | LC diagnosis | AUC 0.73 |
| Ajona 2018 [51] | n = 49 (M 40) Type: AC 12, SQCC 22, SCC 8, other 7 | n = 22 (M 14) | Cell-free BALF supernatant | ELISA | Complement C4d | LC diagnosis | AUC 0.80 |
| Li 2013 [52] | n = 18 (M 7) Age: 51–83 y Type: AC | n = 6 (M 3) Age: 18–85 y | Cell-free proteins from BALF supernatant | ELISA | Napsin A >55 ng/mg total protein | LC diagnosis | AUC 0.85 Sens 84% Spec 67% |
| Uribarri 2014 [53] | n = 204 (M 177) Mean age: 63.0 ± 10.7 y Type: AC 59, SQCC 80, SCC 63, other 2 Stage: I 14, II 4, III 60, IV 109, undefined 17 | n = 48 (M 38) Mean age: 54.9 ± 14.0 y | Cell-free proteins from BALF supernatant | Fluorescent bead-based immunoassay | 5-protein (APOA1, CO4A, CRP, GSTP1, SAMP) prediction model | LC diagnosis | AUC 0.94 Sens 95% Spec 81% |
| 2-protein (STMN1, GSTP1) prediction model | Discrimination of SCC from NSCLC | AUC 0.80 Sens 90% Spec 57% | |||||
| Ortea 2016 [54] | n = 12 (M 8) Median age: 64 y Type: AC Stage: I-II 2, III-IV 10 | n = 10 (M 10) Median age: 61 | Cell-free proteins from BALF supernatant | Liquid chromatography–mass spectrometry | Discriminant analysis of a 44-protein panel | LC diagnosis | Sens 92% Spec 70% |
| Almatroodi 2015 [55] | n = 8 (M 5) Mean age: 68.1 ± 7.6 y Type: AC Stage: I 2, II 2, III 1, IV 3 | n = 8 (M 3) Mean age: 60 ± 8.7 y | Cellular proteins from BALF cell pellets | Liquid chromatography–mass spectrometry | 33 upregulated proteins | LC diagnosis | - |
| Carvalho 2017 [56] | n = 49 Type: AC 28, SQCC 10, SCC 4, LCC 1, other 6 | n = 41 | Cell-free proteins from BALF supernatant | Liquid chromatography–mass spectrometry | Different spectral count values from all abundant proteins | LC diagnosis | - |
| 133 differentially expressed proteins | - | ||||||
| Hmmier 2017 [57] | n = 26 (M 13) Mean age: 65 y Type: AC 13, SQCC 13 Stage: I-II 15, III-IV 11 | n = 16 (M 8) Mean age: 56 y | Cell-free proteins from BALF supernatant | Liquid chromatography–mass spectrometry | 267 differentially expressed proteins | AC diagnosis | - |
| 261 differentially expressed proteins | SQCC diagnosis | - | |||||
| 292 differentially expressed proteins | Discrimination of SQCC from AC | - | |||||
| Liu 2021 [58] | n = 85 (M 60) Type: AC 32, SQCC 32, SCC 21 Stage: I-II 30, III-IV 42, unknown 13 | n = 33 (M 20) | Cell-free proteins from BALF supernatant | Lectin microarray | 3-lectin (ECA, GSL-I, RCA120) prediction model | LC diagnosis | AUC 0.96 Sens 92% Spec 94% |
| 4-lectin (DBA, STL, UEA-I, BPL) prediction model | Discrimination of AC from other subtypes | AUC 0.62 Sens 71% Spec 59% | |||||
| 1-lectin (PNA) prediction model | Discrimination of AC from other subtypes | AUC 0.69 Sens 80% Spec 67% | |||||
| 6-lectin (STL, BS-I, PTL-II, SBA, PSA, GNA) prediction model | Discrimination of AC from other subtypes | AUC 0.72 Sens 72% Spec 68% | |||||
| 6-lectin (MAL-II, LTL, GSL-I, RCA120, PTL-II, PWM) prediction model | Discrimination of early from advanced stage | AUC 0.86 Sens 83% Spec 81% | |||||
| Kwiecien 2017 [60] | n = 18 (M 12) Age: 50–81 y Type: AC 4, SQCC 9, NSCLC NOS 4 Stage: I 4, II 11, III 3 | - | Immune cells from BALF cell pellets | Antibody-specific flow cytometry | % Tregs, CTLA-4+ Tregs (↑) | LC diagnosis (affected vs. healthy lung) | - |
| Hu 2019 [63] | n = 52 (M 29) Age: 39–73 y Type: NSCLC 26, SCC 26 | n = 20 (M 12) Age: 35–75 y | Immune cells from BALF cell pellets | Antibody-specific flow cytometry | % PD-1+ Tph (↓), PD-1+ Tfh/Tph (↑) | SCC diagnosis | - |
| Hu 2020 [64] | n = 67 (M 46) Age: 39–75 y Type: AC 18, SQCC 17, SCC 32 Stage: 0–IIIA 39, IIIB-IV 28 | n = 14 (M 10) Age: 33–71 y | Immune cells from BALF cell pellets | Antibody-specific flow cytometry | Tregs (↑) | LC diagnosis Discrimination of SCC from NSCLC Discrimination of advanced SCC | - |
| IL-10+ CD206+ CD14+ M2-like macrophages (↑) | LC diagnosis Discrimination of SCC from NSCLC Discrimination of advanced SCC Overall survival (worse) | - | |||||
| Cell-free BALF supernatant | Cytometric bead array | IL-10 (↑) | LC diagnosis Discrimination of SCC from NSCLC Discrimination of advanced SCC Overall survival (worse) | - | |||
| Masuhiro 2022 [65] | n = 12 (M 9) Age: 55–70 y Type: AC 7 | - | Cell-free BALF supernatant | Cytometric bead array | CXCL9 (↑) | Prediction of treatment response (immunotherapy) | - |
| Bacterial DNA from BALF supernatant | 16S rRNA sequencing | Bacterial alpha diversity (↑), Proteobacteria (↓), Bacteroidetes (↑) | - | ||||
| n = 7 | Cellular RNA from BALF cell pellets | RNA sequencing | 87 genes were upregulated and 28 were downregulated | - | |||
| Zhong 2021 [70] | n = 12 | n = 6 | Tumor cells from BALF cell pellets | Antibody-specific immunostaining + fluorescence in situ hybridization | Circulating tumor cell count ≥2 | LC diagnosis | Sens 75% Spec 100% |
| Schmid 2015 [72] | n = 26 (M 16) Mean age: 60.2 ± 8.3 y Type: AC 20, SQCC 6 | n = 21 (M 13) Mean age: 64.7 ± 8.4 y | Cell-free BALF supernatant | BLEIA | ATP, ADP (↑) | LC diagnosis | - |
| Cellular RNA from BALF cell pellets | mRNA-specific reverse transcription-PCR | CD39 (↑) | LC diagnosis Discrimination of metastatic disease | - | |||
| P2X4, P2X7, P2Y1 (↑) | Discrimination of metastatic disease | - | |||||
| Callejón-Leblic 2016 [73] | n = 24 (M 16) Mean age: 66 ± 11 y | n = 31 (M 23) Mean age: 56 ± 13 y | Cell-free metabolites from BALF supernatant | Direct infusion mass spectrometry | Carnitine | LC diagnosis | AUC 0.88 |
| Adenine | AUC 0.83 | ||||||
| Choline | AUC 0.78 | ||||||
| Gas chromatography–mass spectrometry | Glycerol | AUC 0.89 | |||||
| Phosphoric acid | AUC 0.79 | ||||||
| Callejón-Leblic 2018 [75] | n = 24 (M 20) Mean age: 65 ± 13 y Type: NSCLC 22, SCC 2 | n = 31 (M 27) Mean age: 54 ± 14 y | Cell-free elements from BALF supernatant | Inductive coupled plasma mass spectrometry | Mn | LC diagnosis | AUC 0.75 |
| V/Cu ratio | AUC 0.76 | ||||||
| Suresh 2019 [94] | n = 18 (M 13) Type: NSCLC 15, other 3 | - | Immune cells from BALF cell pellets | Antibody-specific flow cytometry | % PD-1hi/CTLA-4hi Tregs (↓), % IL-1RA-expressing B cells (↓), % central memory T cells (↑), % CD8+ TNF-αhi T cells (↑), % IL-1βhi monocytes (↑) | Prediction of CIP development | - |
| Cell-free BALF supernatant | V-plex immunoassays | IL-1β (↓), IL-8 (↓), MIP-3α (↓), IL-12p40 (↑), IP-10 (↑) | - | ||||
| Crohns 2010 [104] | n = 36 (M 29) Age: 47–82 y Type: AC 1, SQCC 33, SCC 2 Stage: I 1, III 18, IV 17 | n = 36 (M 16) Age: 18–75 y | Cell-free BALF supernatant | ELISA | Il-6 (↑) | LC diagnosis | - |
| IL-8 (↑) | Overall survival (worse) | - | |||||
| Yamagishi 2017 [105] | n = 22 (M 16) Type: AC 8, SQCC 8, SCC 4, unknown 2 | - | Cell-free BALF supernatant | ELISA | MMP-9 (↑) | Prediction of radiation pneumonitis | - |
| VEGF (↓) | - | ||||||
| Metagenomic biomarkers | |||||||
| Wang 2019 [76] | n = 51 (M 31) Type: AC 18, SQCC 19, SCC 14 | n = 15 (M 8) | Bacterial DNA from BALF cell pellet | 16S rRNA sequencing | Microbial diversity (↓) | LC diagnosis | - |
| Treponema | AUC 0.86 | ||||||
| Cheng 2020 [77] | n = 32 (M 23) Mean age: 64.3 ± 8.4 y Type: AC 16, SQCC 9, SCC 7 Stage: I 7, III 6, IV 19 | n = 22 (M 12) Mean age: 56.5 ± 14.3 y | Bacterial DNA from BALF supernatant | 16S rRNA sequencing | 10-genera (f:Pseudomonadaceae, Capnocytophaga, Stenotrophomonas, Microbacterium, Gemmiger, c:TM7-3, Oscillospira, Blautia, Lautropia, Sediminibacterium) prediction model | LC diagnosis | AUC 0.79 |
| Lee 2016 [78] | n = 20 (M 13) Median age: 64 y Type: AC 13, SQCC 5, SCC 2 Stage: II 6, III 8, IV 6 | n = 8 (M 7) Median age: 58.5 y | Bacterial DNA from BALF supernatant | 16S rRNA pyrosequencing | Veillonella | LC diagnosis | AUC 0.86 |
| Megasphaera | AUC 0.78 | ||||||
| Combined panel | AUC 0.89 | ||||||
| Patnaik 2021 [79] | n = 36 (M 16) Type: AC 24, SQCC 11, other 1 Stage: I | - | Bacterial DNA from BALF supernatant | 16S rRNA sequencing | 19-genera microbiome signature | Prediction of recurrence after surgery | AUC 0.77 |
| Zheng 2021 [80] | n = 32 Type: NSCLC Stage: I-II 23, III-IV 9 | n = 15 | Bacterial DNA from BALF supernatant | 16S rRNA sequencing | Differentiated abundance of 19 species | LC diagnosis | - |
| Jang 2021 [81] | n = 11 (M 9) Median age: 63 y Type: AC 8, SQCC 3 Stage: III 5, IV 6 | - | Bacterial DNA from BALF supernatant | 16S rRNA sequencing | Haemophilus influenzae (↓), Neisseria perflava (↓), Veillonella dispar (↑) | Prediction of treatment response (immunotherapy) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalkanis, A.; Papadopoulos, D.; Testelmans, D.; Kopitopoulou, A.; Boeykens, E.; Wauters, E. Bronchoalveolar Lavage Fluid-Isolated Biomarkers for the Diagnostic and Prognostic Assessment of Lung Cancer. Diagnostics 2022, 12, 2949. https://doi.org/10.3390/diagnostics12122949
Kalkanis A, Papadopoulos D, Testelmans D, Kopitopoulou A, Boeykens E, Wauters E. Bronchoalveolar Lavage Fluid-Isolated Biomarkers for the Diagnostic and Prognostic Assessment of Lung Cancer. Diagnostics. 2022; 12(12):2949. https://doi.org/10.3390/diagnostics12122949
Chicago/Turabian StyleKalkanis, Alexandros, Dimitrios Papadopoulos, Dries Testelmans, Alexandra Kopitopoulou, Eva Boeykens, and Els Wauters. 2022. "Bronchoalveolar Lavage Fluid-Isolated Biomarkers for the Diagnostic and Prognostic Assessment of Lung Cancer" Diagnostics 12, no. 12: 2949. https://doi.org/10.3390/diagnostics12122949
APA StyleKalkanis, A., Papadopoulos, D., Testelmans, D., Kopitopoulou, A., Boeykens, E., & Wauters, E. (2022). Bronchoalveolar Lavage Fluid-Isolated Biomarkers for the Diagnostic and Prognostic Assessment of Lung Cancer. Diagnostics, 12(12), 2949. https://doi.org/10.3390/diagnostics12122949








