Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets and Preprocessing
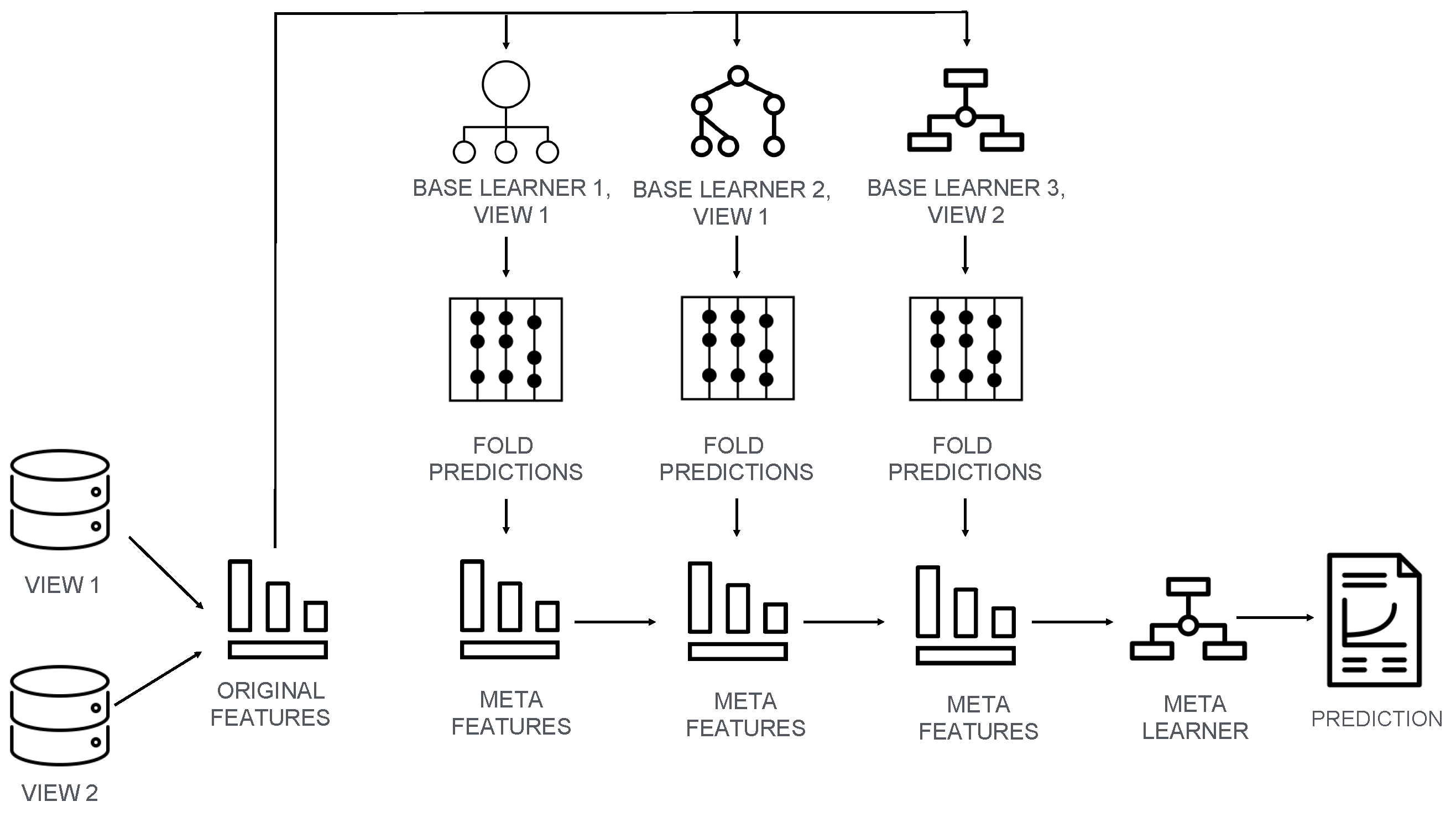
2.2. Machine Learning Model and Training Procedure
2.3. Performance Metrics and Handling Class Imbalance
2.4. Model Interpretability
3. Results
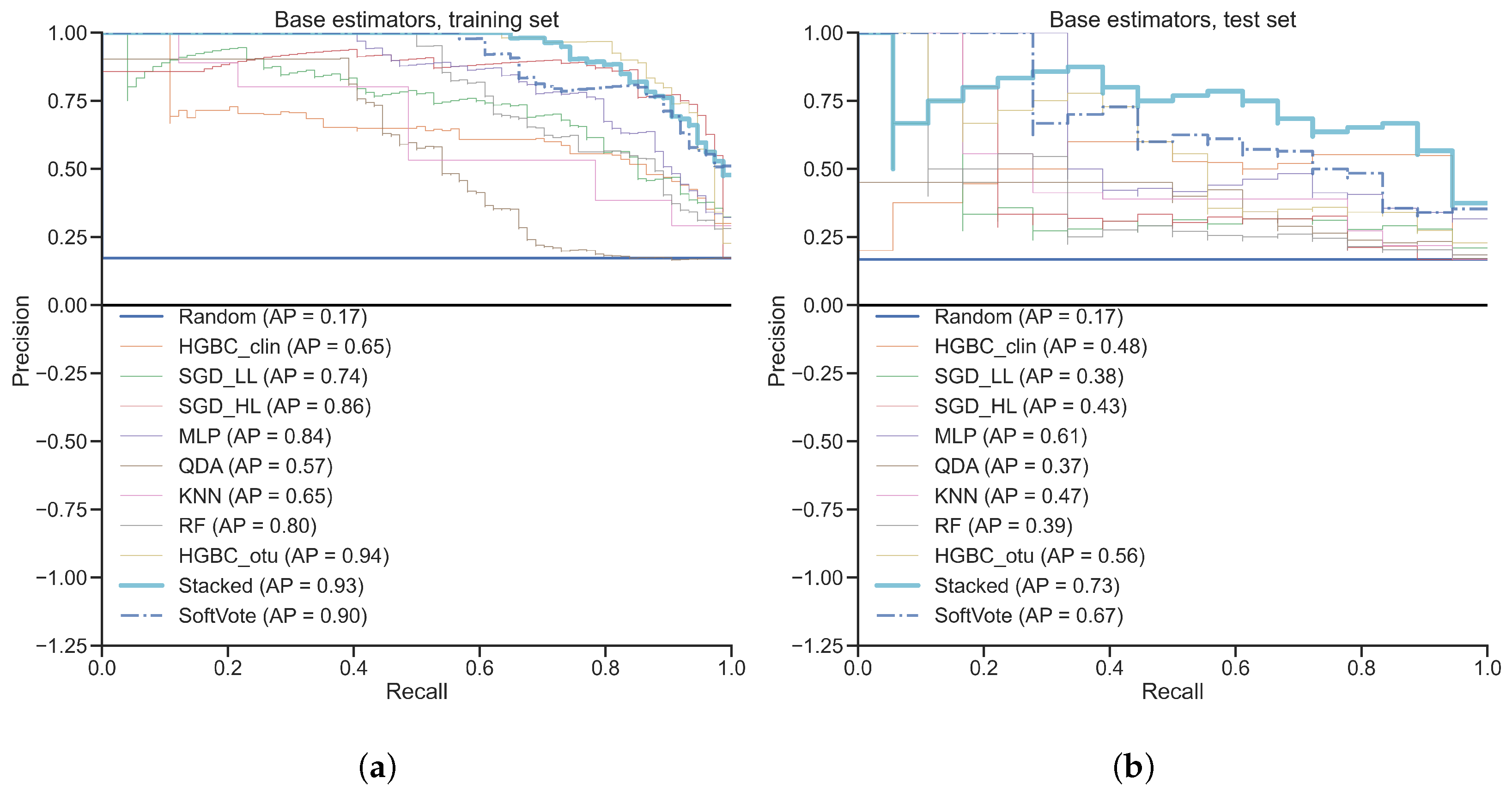
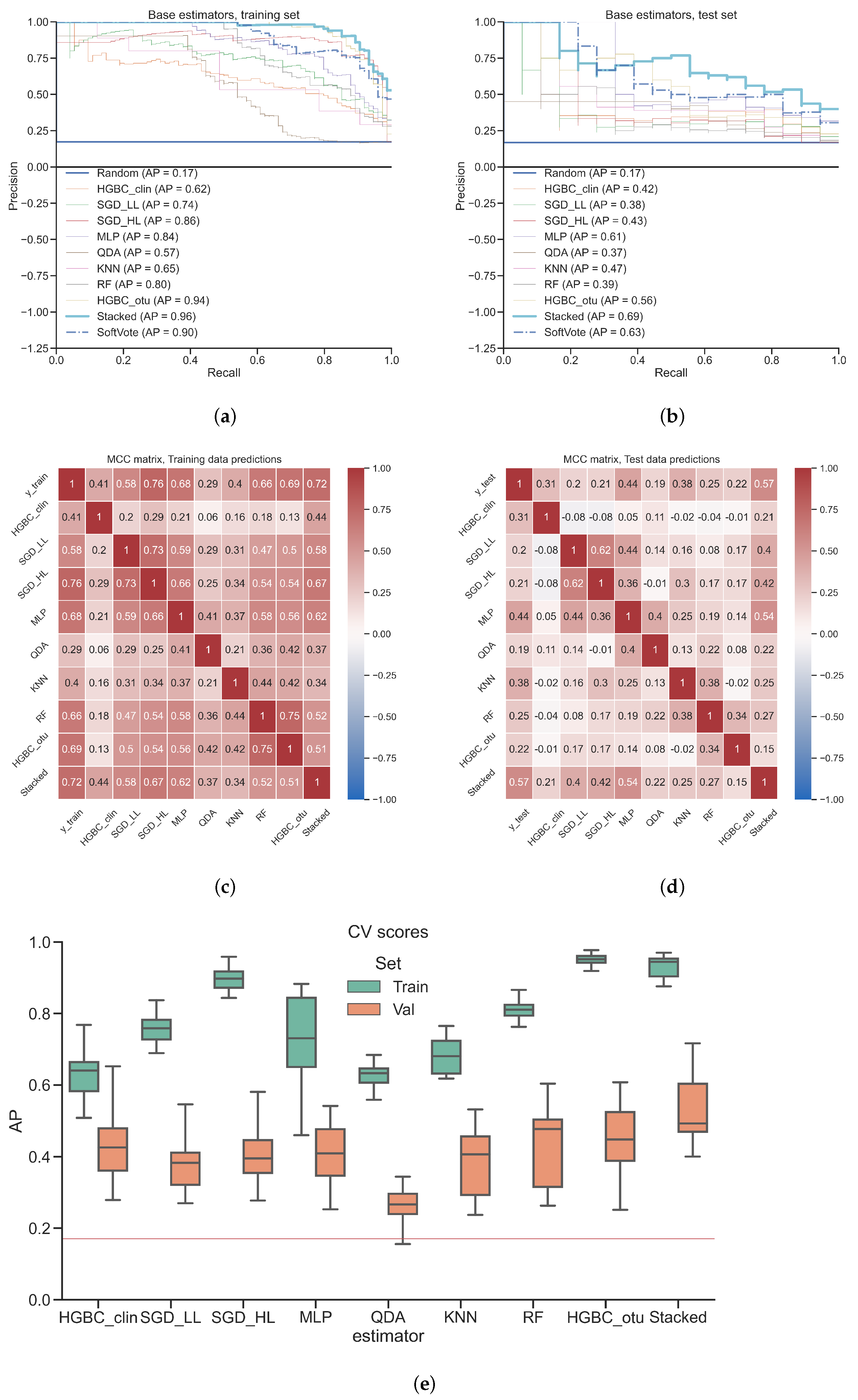
3.1. Classification of Gut Microbiota from Inflammatory Bowel Disease Patients
3.2. Classification of Gut Microbiota from Colorectal Cancer Patients
4. Discussion
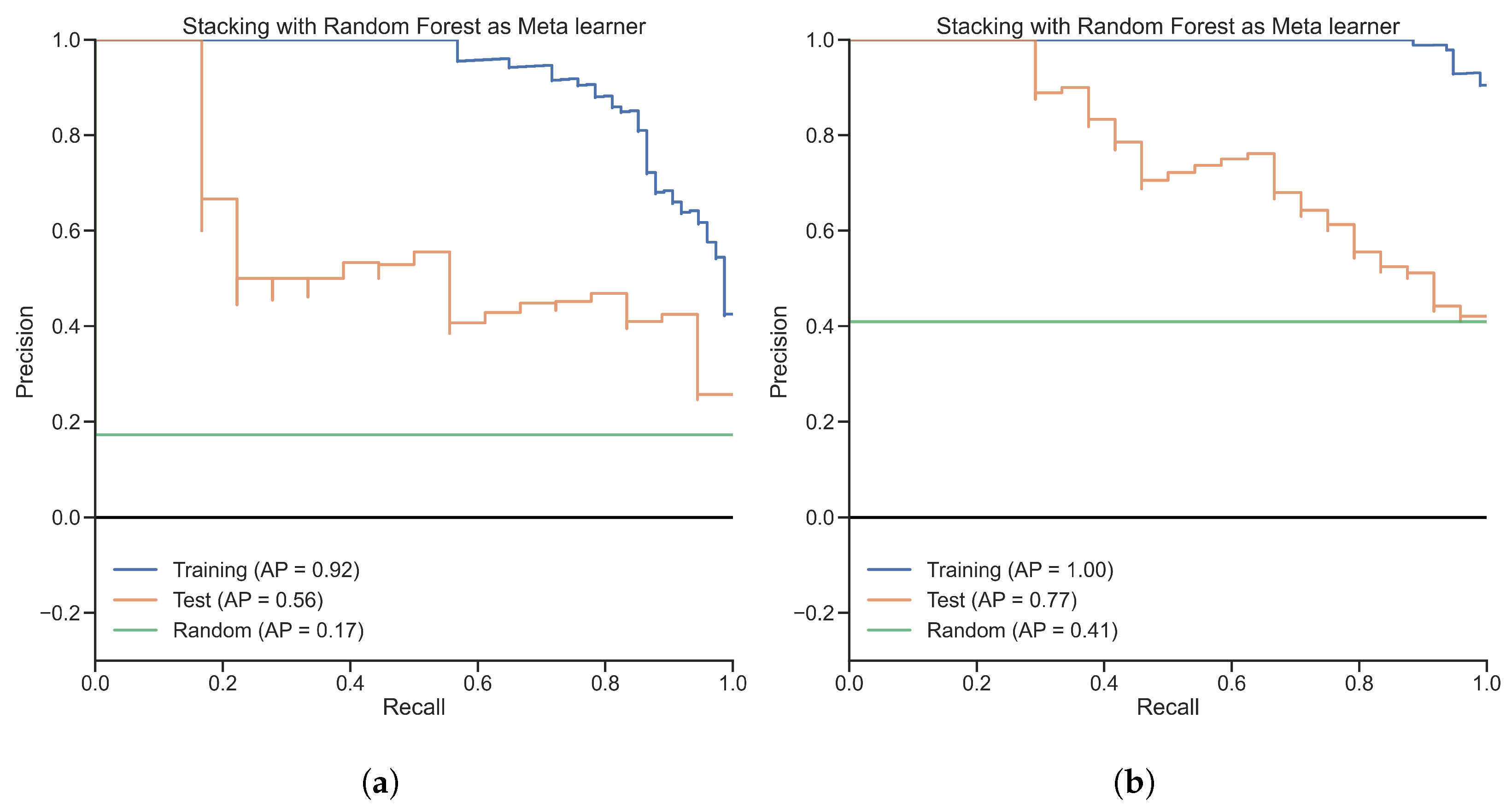
4.1. Stacking as a More Powerful Ensemble Method than Random Forest
4.2. Meta Learner’s Role in Stacking
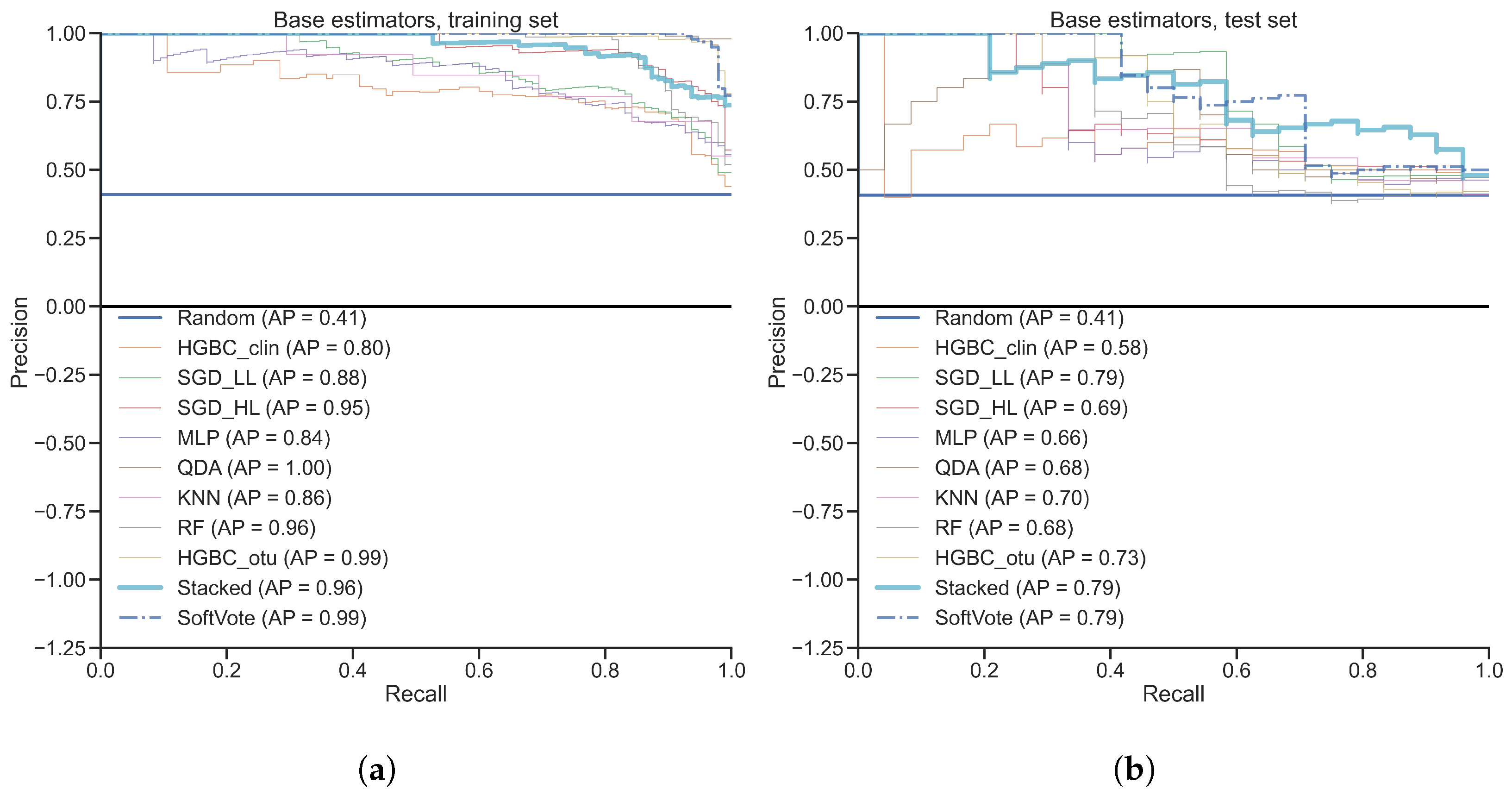
4.3. Diversity of Base Learner’s Role in Stacking
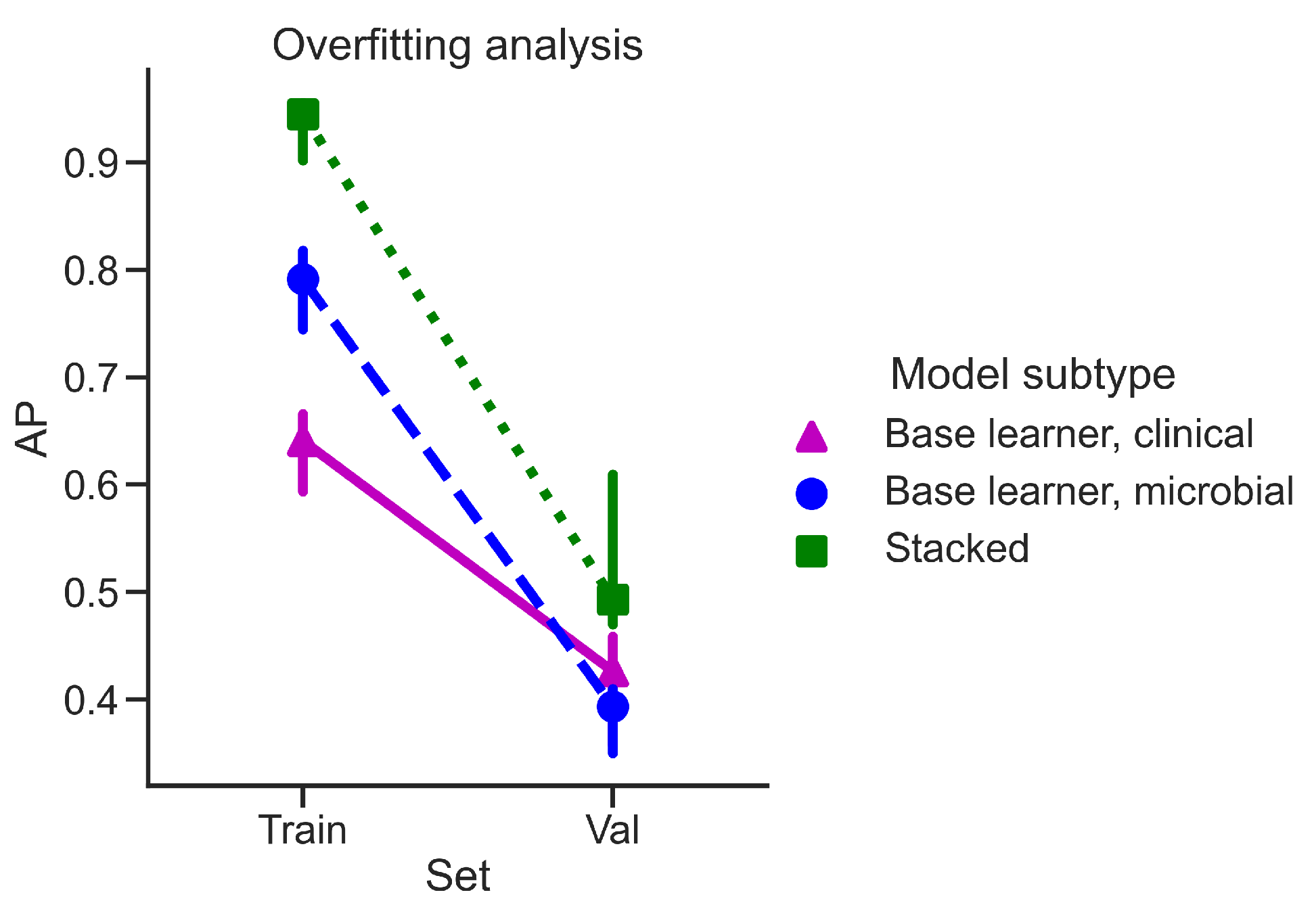
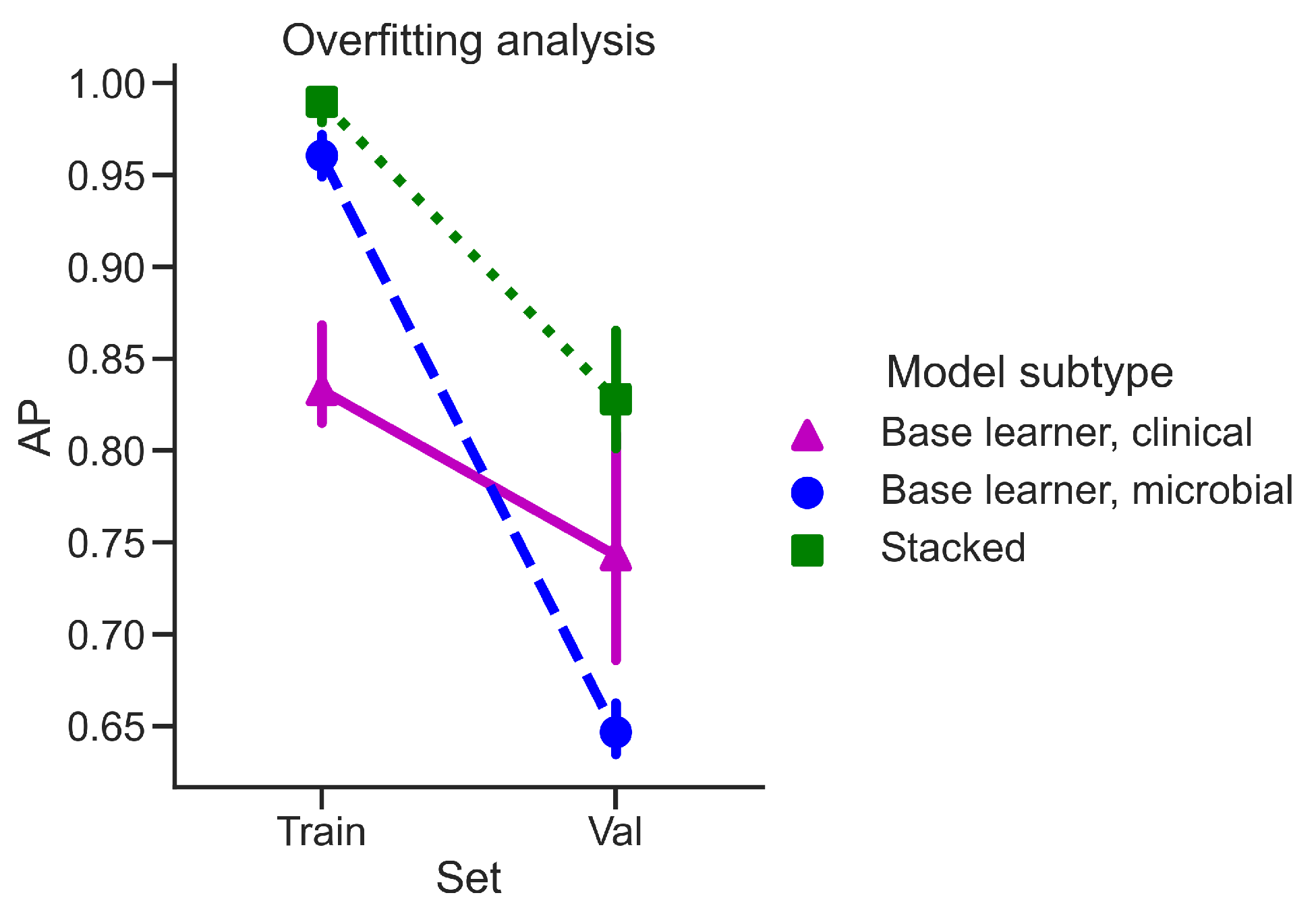
4.4. Overfitting Analysis
4.5. Model Interpretation and Further Examples of Stacking
4.6. Study Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Average Precision |
| AUROC | Area Under Receiver Operating Characteristic |
| MCC | Matthews’s Correlation Coefficient |
| SGD_LL | Stochastic Gradient Descent classifier with Logistic Loss |
| SGD_HL | Stochastic Gradient Descent classifier with modified Huber Loss |
| KNN | K-nearest Neighbors Classifier |
| MLP | Multi-layer Perceptron |
| QDA | Quadratic Discriminant Analysis |
| RF | Random Forest |
| HGBC | Histogram-based Gradient Boosting Classification |
Appendix A

| Feature Name | Type | Reason |
|---|---|---|
| diagnosis | Categorical | Training label |
| external_id | Categorical | Irrelevant |
| run_prefix | Categorical | Irrelevant |
| body_site | Categorical | Training label leakage |
| diseasesubtype | Categorical | Explicit training label leakage |
| gastrointest_disord | Categorical | Explicit training label leakage |
| host_subject_id | Categorical | Irrelevant |
| disease_stat | Categorical | Implicit training label leakage |
| disease_extent | Categorical | Implicit training label leakage |
| ileal_invovlement | Categorical | Implicit training label leakage |
| gastric_involvement | Categorical | Implicit training label leakage |
| antibiotics | Categorical | Irrelevant |
| steroids | Categorical | Irrelevant |
| collection | Categorical | Irrelevant |
| biologics | Categorical | Irrelevant |
| birthdate | Categorical | Irrelevant |
| body_habitat | Categorical | Implicit training label leakage |
| body_product | Categorical | Irrelevant |
| disease_duration | Numerical | Irrelevant |
| immunosup | Categorical | Irrelevant |
| mesalamine | Categorical | Irrelevant |
| sample_type | Categorical | Irrelevant |
| smoking | Categorical | Irrelevant |
| Feature Name | Type | Description |
|---|---|---|
| Age | Numerical | Age in years |
| b_cat | Categorical | Montreal classification of inflammatory bowel disease |
| biopsy_location | Categorical | Location of biopsy |
| inflammationstatus | Categorical | Inflammation status |
| perianal | Categorical | Extension to anal area |
| race | Categorical | Race |
| sex | Categorical | Biological gender |
| Feature Name | Type | Reason |
|---|---|---|
| Dx_Bin | Categorical | Training label |
| Site | Categorical | Irrelevant |
| Location | Categorical | Irrelevant |
| Ethnic | Categorical | Irrelevant |
| fit_result | Numerical | Training label leakage |
| Hx_Prev | Categorical | Training label leakage |
| Hx_Fam_CRC | Categorical | Training label leakage |
| Hx_of_Polyps | Categorical | Training label leakage |
| stage | Categorical | Training label leakage |
| Gender | Categorical | Encoded as “Sex” |
| White | Categorical | Encoded as “Ethnicity” |
| Native | Categorical | Encoded as “Ethnicity” |
| Black | Categorical | Encoded as “Ethnicity” |
| Pacific | Categorical | Encoded as “Ethnicity” |
| Asian | Categorical | Encoded as “Ethnicity” |
| Other | Categorical | Encoded as “Ethnicity” |
| Weight | Numerical | Encoded as “BMI” |
| Height | Numerical | Encoded as “BMI” |
| Feature Name | Type | Description |
|---|---|---|
| BMI | Numerical | Body Mass Index |
| Age | Numerical | Age in years |
| Smoke | Categorical | Smoking |
| Diabetic | Categorical | Diabetes Melitus |
| NSAID | Categorical | Anti-inflammatory medication |
| Diabetes_Med | Categorical | Anti-diabetes medication |
| Abx | Categorical | Antibiotic medication |
| Ethnicity | Categorical | Race |
| Sex | Categorical | Biological gender |
References
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Lv, G.; Cheng, N.; Wang, H. The gut microbiota, tumorigenesis, and liver diseases. Engineering 2017, 3, 110–114. [Google Scholar] [CrossRef]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases—Does a common dysbiosis exist? Microbiome 2018, 6, 1–15. [Google Scholar] [CrossRef]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic review: The gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Alexander, K.L.; Zhao, Q.; Reif, M.; Rosenberg, A.F.; Mannon, P.J.; Duck, L.W.; Elson, C.O. Human microbiota flagellins drive adaptive immune responses in Crohn’s disease. Gastroenterology 2021, 161, 522–535. [Google Scholar] [CrossRef]
- Ghannam, R.B.; Techtmann, S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021, 19, 1092–1107. [Google Scholar] [CrossRef]
- Sudhakar, P.; Machiels, K.; Verstockt, B.; Korcsmaros, T.; Vermeire, S. Computational biology and machine learning approaches to understand mechanistic microbiome-host interactions. Front. Microbiol. 2021, 12, 618856. [Google Scholar] [CrossRef]
- Douglas, G.M.; Hansen, R.; Jones, C.; Dunn, K.A.; Comeau, A.M.; Bielawski, J.P.; Tayler, R.; El-Omar, E.M.; Russell, R.K.; Hold, G.L.; et al. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome 2018, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Heshiki, Y.; Vazquez-Uribe, R.; Li, J.; Ni, Y.; Quainoo, S.; Imamovic, L.; Li, J.; Sørensen, M.; Chow, B.K.; Weiss, G.J.; et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Vilas-Boas, F.; Ribeiro, T.; Afonso, J.; Cardoso, H.; Lopes, S.; Moutinho-Ribeiro, P.; Ferreira, J.; Mascarenhas-Saraiva, M.; Macedo, G. Deep Learning for Automatic Differentiation of Mucinous versus Non-Mucinous Pancreatic Cystic Lesions: A Pilot Study. Diagnostics 2022, 12, 2041. [Google Scholar] [CrossRef]
- Mascarenhas, M.; Afonso, J.; Ribeiro, T.; Cardoso, H.; Andrade, P.; Ferreira, J.P.; Saraiva, M.M.; Macedo, G. Performance of a deep learning system for automatic diagnosis of protruding lesions in colon capsule endoscopy. Diagnostics 2022, 12, 1445. [Google Scholar] [CrossRef]
- Nogueira-Rodríguez, A.; Reboiro-Jato, M.; Glez-Peña, D.; López-Fernández, H. Performance of Convolutional Neural Networks for Polyp Localization on Public Colonoscopy Image Datasets. Diagnostics 2022, 12, 898. [Google Scholar] [CrossRef]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine learning meta-analysis of large metagenomic datasets: Tools and biological insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef]
- Topçuoğlu, B.D.; Lesniak, N.A.; Ruffin IV, M.T.; Wiens, J.; Schloss, P.D. A framework for effective application of machine learning to microbiome-based classification problems. MBio 2020, 11, e00434-20. [Google Scholar] [CrossRef]
- Bourel, M.; Segura, A. Multiclass classification methods in ecology. Ecol. Indic. 2018, 85, 1012–1021. [Google Scholar] [CrossRef]
- Statnikov, A.; Henaff, M.; Narendra, V.; Konganti, K.; Li, Z.; Yang, L.; Pei, Z.; Blaser, M.J.; Aliferis, C.F.; Alekseyenko, A.V. A comprehensive evaluation of multicategory classification methods for microbiomic data. Microbiome 2013, 1, 1–12. [Google Scholar] [CrossRef]
- Caruana, R.; Lou, Y.; Gehrke, J.; Koch, P.; Sturm, M.; Elhadad, N. Intelligible models for healthcare: Predicting pneumonia risk and hospital 30-day readmission. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, Australia, 10–13 August 2015; pp. 1721–1730. [Google Scholar]
- Nauta, M.; Walsh, R.; Dubowski, A.; Seifert, C. Uncovering and correcting shortcut learning in machine learning models for skin cancer diagnosis. Diagnostics 2021, 12, 40. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Lou, Y.; Caruana, R.; Gehrke, J.; Hooker, G. Accurate intelligible models with pairwise interactions. In Proceedings of the 19th ACM SIGKDD international conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 623–631. [Google Scholar]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Džeroski, S.; Ženko, B. Is combining classifiers with stacking better than selecting the best one? Mach. Learn. 2004, 54, 255–273. [Google Scholar] [CrossRef]
- Sesmero, M.P.; Ledezma, A.I.; Sanchis, A. Generating ensembles of heterogeneous classifiers using stacked generalization. WIley Interdiscip. Rev. Data Min. Knowl. Discov. 2015, 5, 21–34. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.; Lu, W.; Wu, T.; Yuan, W.; Zhu, J.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. Human gut microbiome aging clocks based on taxonomic and functional signatures through multi-view learning. Gut Microbes 2022, 14, 2025016. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Knights, D.; Kostic, A.D.; Knight, R.; Xavier, R.J. A microbiome foundation for the study of Crohn’s disease. Cell Host Microbe 2017, 21, 301–304. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Baxter, N.T.; Ruffin, M.T.; Rogers, M.A.; Schloss, P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016, 8, 1–10. [Google Scholar] [CrossRef]
- Battaglia, T. A Repository for Large-Scale Microbiome Datasets. 2022. Available online: https://github.com/twbattaglia/MicrobeDS (accessed on 13 October 2022).
- The Laboratory of Pat Schloss at the University of Michigan. 2022. Available online: https://github.com/SchlossLab/Baxter_glne007Modeling_GenomeMed_2015 (accessed on 13 October 2022).
- Yeo, I.K.; Johnson, R.A. A new family of power transformations to improve normality or symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Branco, P.; Torgo, L.; Ribeiro, R.P. A survey of predictive modeling on imbalanced domains. ACM Comput. Surv. Csur 2016, 49, 1–50. [Google Scholar] [CrossRef]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Yuan, Y.; Zhu, M. A relationship between the average precision and the area under the ROC curve. In Proceedings of the 2015 International Conference on The Theory of Information Retrieval, Northampton, MA, USA, 27–30 September 2015; pp. 349–352. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zadrozny, B.; Langford, J.; Abe, N. Cost-sensitive learning by cost-proportionate example weighting. In Proceedings of the Third IEEE International Conference on Data Mining, Melbourne, FL, USA, 22 November 2003; pp. 435–442. [Google Scholar]
- Chang, C.C.; Huang, T.H.; Shueng, P.W.; Chen, S.H.; Chen, C.C.; Lu, C.J.; Tseng, Y.J. Developing a Stacked Ensemble-Based Classification Scheme to Predict Second Primary Cancers in Head and Neck Cancer Survivors. Int. J. Environ. Res. Public Health 2021, 18, 12499. [Google Scholar] [CrossRef]
- Ting, K.M.; Witten, I.H. Issues in stacked generalization. J. Artif. Intell. Res. 1999, 10, 271–289. [Google Scholar] [CrossRef]
- Ghaemi, M.S.; DiGiulio, D.B.; Contrepois, K.; Callahan, B.; Ngo, T.T.; Lee-McMullen, B.; Lehallier, B.; Robaczewska, A.; Mcilwain, D.; Rosenberg-Hasson, Y.; et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 2019, 35, 95–103. [Google Scholar] [CrossRef]
- Klang, E.; Freeman, R.; Levin, M.A.; Soffer, S.; Barash, Y.; Lahat, A. Machine Learning Model for Outcome Prediction of Patients Suffering from Acute Diverticulitis Arriving at the Emergency Department—A Proof of Concept Study. Diagnostics 2021, 11, 2102. [Google Scholar] [CrossRef]
- Baumgart, D.C. The diagnosis and treatment of Crohn’s disease and ulcerative colitis. Dtsch. ÄRzteblatt Int. 2009, 106, 123. [Google Scholar] [CrossRef]
- Sartor, R.B. Mechanisms of disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 390–407. [Google Scholar] [CrossRef]
- Silva, M.; Pratas, D.; Pinho, A.J. AC2: An Efficient Protein Sequence Compression Tool Using Artificial Neural Networks and Cache-Hash Models. Entropy 2021, 23, 530. [Google Scholar] [CrossRef]
- Janitza, S.; Hornung, R. On the overestimation of random forest’s out-of-bag error. PloS ONE 2018, 13, e0201904. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, B.; He, T.; Li, G.; Jiang, X. Robust biomarker discovery for microbiome-wide association studies. Methods 2020, 173, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Bakir-Gungor, B.; Hacılar, H.; Jabeer, A.; Nalbantoglu, O.U.; Aran, O.; Yousef, M. Inflammatory bowel disease biomarkers of human gut microbiota selected via different feature selection methods. PeerJ 2022, 10, e13205. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Paterson, A.D.; Xu, W. TaxoNN: Ensemble of neural networks on stratified microbiome data for disease prediction. Bioinformatics 2020, 36, 4544–4550. [Google Scholar] [CrossRef]
- Mulenga, M.; Kareem, S.A.; Sabri, A.Q.M.; Seera, M. Stacking and chaining of normalization methods in deep learning-based classification of colorectal cancer using gut microbiome data. IEEE Access 2021, 9, 97296–97319. [Google Scholar] [CrossRef]



| Clinical View Features | Microbial View Features | |||
|---|---|---|---|---|
| # Numerical | # Categorical | # Total | # Unique Genera | |
| Inflammatory bowel disease | 1 | 6 | 6737 | 533 |
| Colorectal cancer | 2 | 7 | 5982 | 239 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imangaliyev, S.; Schlötterer, J.; Meyer, F.; Seifert, C. Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data. Diagnostics 2022, 12, 2514. https://doi.org/10.3390/diagnostics12102514
Imangaliyev S, Schlötterer J, Meyer F, Seifert C. Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data. Diagnostics. 2022; 12(10):2514. https://doi.org/10.3390/diagnostics12102514
Chicago/Turabian StyleImangaliyev, Sultan, Jörg Schlötterer, Folker Meyer, and Christin Seifert. 2022. "Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data" Diagnostics 12, no. 10: 2514. https://doi.org/10.3390/diagnostics12102514
APA StyleImangaliyev, S., Schlötterer, J., Meyer, F., & Seifert, C. (2022). Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data. Diagnostics, 12(10), 2514. https://doi.org/10.3390/diagnostics12102514






