Genome-Wide Association Study Identifies Genetic Variants Associated with Rotator Cuff Tear—A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Population
2.3. Genomic DNA Extraction and Exome Sequencing
2.4. Read Mapping and Variant Analysis
2.5. Variant Filtering
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Longo, U.G.; Berton, A.; Papapietro, N.; Maffulli, N.; Denaro, V. Epidemiology, genetics and biological factors of rotator cuff tears. Rotator Cuff Tear 2012, 57, 1–9. [Google Scholar]
- da Rocha Motta, G.; Amaral, M.V.; Rezende, E.; Pitta, R.; dos Santos Vieira, T.C.; Duarte, M.E.L.; Vieira, A.R.; Casado, P.L. Evidence of genetic variations associated with rotator cuff disease. J. Shoulder Elb. Surg. 2014, 23, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Tashjian, R.Z. Epidemiology, natural history, and indications for treatment of rotator cuff tears. Clin. Sports Med. 2012, 31, 589–604. [Google Scholar] [CrossRef]
- Longo, U.G.; Candela, V.; Berton, A.; Salvatore, G.; Guarnieri, A.; DeAngelis, J.; Nazarian, A.; Denaro, V. Genetic basis of rotator cuff injury: A systematic review. BMC Med. Genet. 2019, 20, 149. [Google Scholar] [CrossRef] [PubMed]
- Benson, R.; McDonnell, S.; Knowles, H.; Rees, J.; Carr, A.; Hulley, P. Tendinopathy and tears of the rotator cuff are associated with hypoxia and apoptosis. J. Bone Jt. Surg. Br. Vol. 2010, 92, 448–453. [Google Scholar] [CrossRef]
- Chaudhury, S.; Carr, A.J. Lessons we can learn from gene expression patterns in rotator cuff tears and tendinopathies. J. Shoulder Elb. Surg. 2012, 21, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.V.; Silverio, L.; Sperling, J.W. Strategies in biologic augmentation of rotator cuff repair: A review. Clin. Orthop. Relat. Res.® 2010, 468, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.; Figueiredo, E.A.; Leal, M.F.; Ejnisman, B. Genetics in Rotator Cuff Tears: First Steps to the Future. In Rotator Cuff Across the Life Span; Springer: Berlin/Heidelberg, Germany, 2019; pp. 43–46. [Google Scholar]
- Maffulli, N.; Longo, U.G.; Berton, A.; Loppini, M.; Denaro, V. Biological factors in the pathogenesis of rotator cuff tears. Sports Med. Arthrosc. Rev. 2011, 19, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Assunção, J.H.; Godoy-Santos, A.L.; Dos Santos, M.C.L.; Malavolta, E.A.; Gracitelli, M.E.; Ferreira Neto, A.A. Matrix metalloproteases 1 and 3 promoter gene polymorphism is associated with rotator cuff tear. Clin. Orthop. Relat. Res.® 2017, 475, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Kluger, R.; Burgstaller, J.; Vogl, C.; Brem, G.; Skultety, M.; Mueller, S. Candidate gene approach identifies six SNPs in tenascin-C (TNC) associated with degenerative rotator cuff tears. J. Orthop. Res. 2017, 35, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Roos, T.R.; Roos, A.K.; Avins, A.L.; Ahmed, M.A.; Kleimeyer, J.P.; Fredericson, M.; Ioannidis, J.P.A.; Dragoo, J.L.; Kim, S.K. Genome-wide association study identifies a locus associated with rotator cuff injury. PLoS ONE 2017, 12, e0189317. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Nicolae, D.L.; Gamazon, E.; Zhang, W.; Duan, S.; Dolan, M.E.; Cox, N.J. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010, 6, e1000888. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Melissa Coon, M.; Tung Nguyen, T.; Luan Wang, L.; Susan, J.; Land, S.J.; Lu, X.; Douglas, M.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.; Raleigh, S.M. Genetic risk factors for musculoskeletal soft tissue injuries. In Genetics and Sports; Karger Publishers: Basel, Switzerland, 2009; pp. 136–149. [Google Scholar]
- Harvie, P.; Ostlere, S.; Teh, J.; McNally, E.G.; Clipsham, K.; Burston, B.J.; Pollard, T.C.B.; Carr, A.J. Genetic influences in the aetiology of tears of the rotator cuff: Sibling risk of a full-thickness tear. J. Bone Jt. Surg. Br. Vol. 2004, 86, 696–700. [Google Scholar] [CrossRef]
- Ramos, M.I.P.; Tian, L.; de Ruiter, E.J.; Song, C.; Paucarmayta, A.; Singh, A.; Elshof, E.; Vijver, S.V.; Shaik, J.; Bosiacki, J.; et al. Cancer immunotherapy by NC410, a LAIR-2 Fc protein blocking human LAIR-collagen interaction. Elife 2021, 10, e62927. [Google Scholar] [CrossRef] [PubMed]
- Camargo, C.M.; Augusto, D.G.; Petzl-Erler, M.L. Differential gene expression levels might explain association of LAIR2 polymorphisms with pemphigus. Hum. Genet. 2016, 135, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Talukder, A.; Meng, Q.; Kumar, R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene 2006, 25, 1311–1319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Y.; Liu, H.; Jin, M.; Zhang, M.; Pan, Y.; Zhang, S.; Li, Q.; Chen, K. The joint association of REST and NFKB1 polymorphisms on the risk of colorectal cancer. Ann. Hum. Genet. 2012, 76, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Mousley, J.J.; Hill-Buxton, L.-M.; Gill, S.D.; McGee, S.L.; Page, R.S. Polymorphisms and alterations in gene expression associated with rotator cuff tear and healing following surgical repair: A systematic review. J. Shoulder Elb. Surg. 2021, 30, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Tashjian, R.Z.; Kim, S.K.; Roche, M.D.; Jones, K.B.; Teerlink, C.C. Genetic variants associated with rotator cuff tearing utilizing multiple population-based genetic resources. J. Shoulder Elb. Surg. 2021, 30, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Bonato, L.L.; Quinelato, V.; Pinheiro, A.; Amaral, M.V.G.; de Souza, F.N.; Lobo, J.C.; Aguiar, D.P.; Augusto, L.M.M.; Vieira, A.R.; Salles, J.I. ESRRB polymorphisms are associated with comorbidity of temporomandibular disorders and rotator cuff disease. Int. J. Oral Maxillofac. Surg. 2016, 45, 323–331. [Google Scholar] [CrossRef]
- Peach, C.A.; Zhang, Y.; Dunford, J.E.; Brown, M.A.; Carr, A.J. Cuff tear arthropathy: Evidence of functional variation in pyrophosphate metabolism genes. Clin. Orthop. Relat. Res.® 2007, 462, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Tashjian, R.Z.; Granger, E.K.; Farnham, J.M.; Cannon-Albright, L.A.; Teerlink, C.C. Genome-wide association study for rotator cuff tears identifies two significant single-nucleotide polymorphisms. J. Shoulder Elb. Surg. 2016, 25, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Teerlink, C.C.; Cannon-Albright, L.A.; Tashjian, R.Z. Significant association of full-thickness rotator cuff tears and estrogen-related receptor-β (ESRRB). J. Shoulder Elb. Surg. 2015, 24, e31–e35. [Google Scholar] [CrossRef] [PubMed]
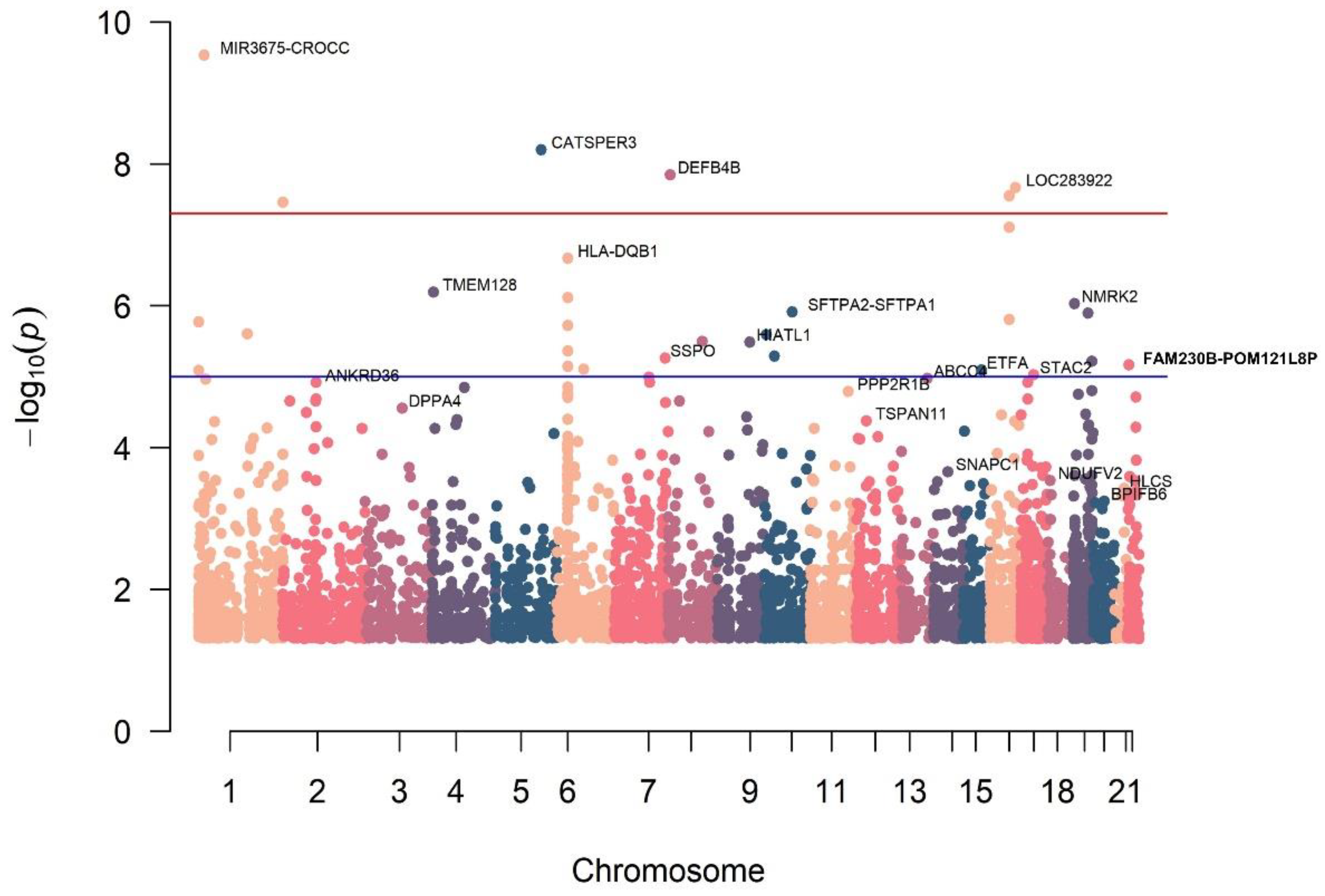
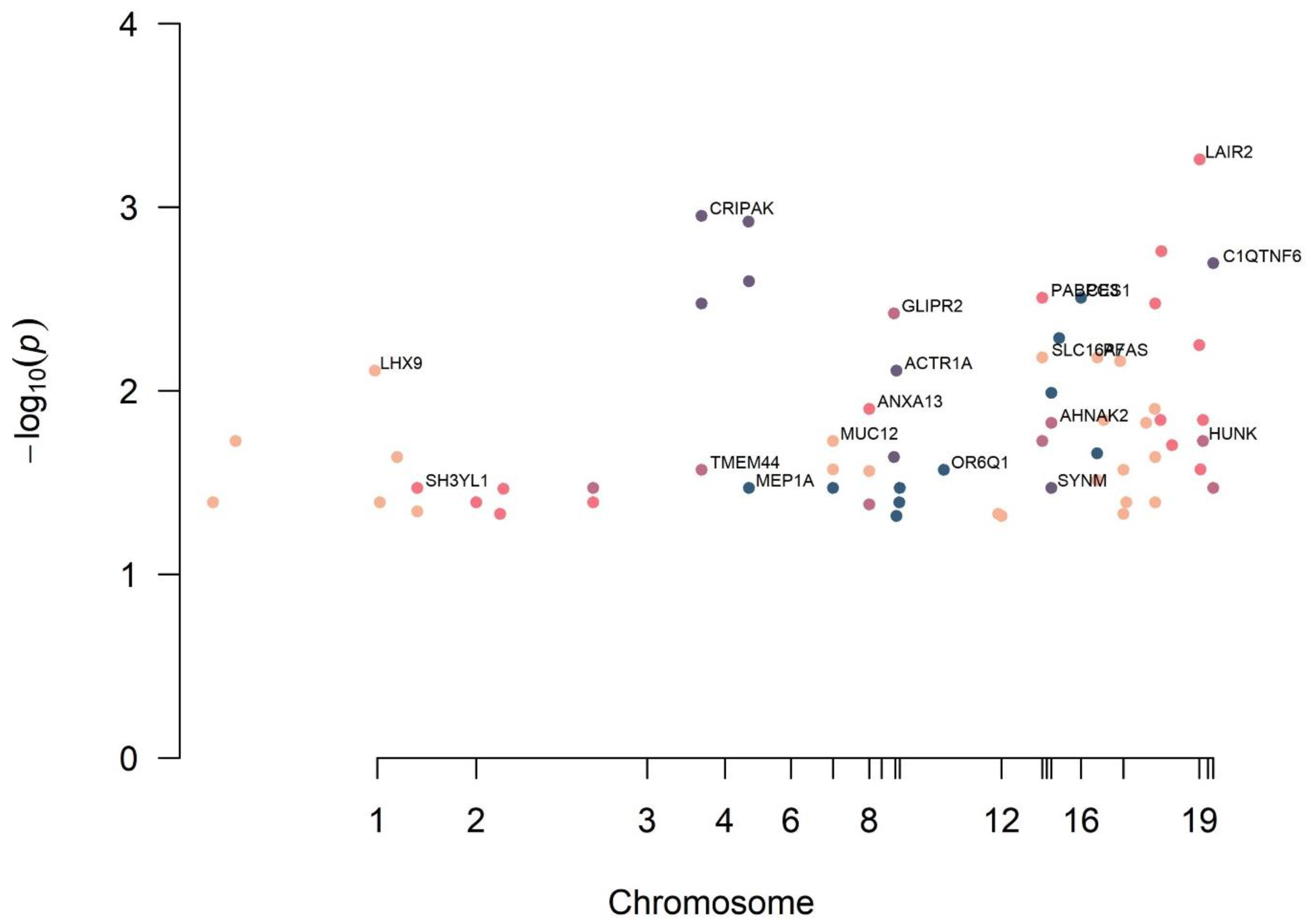
| Variable | RC (n = 20) | Control (n = 20) | OR | p-Value | |
|---|---|---|---|---|---|
| Gender, n (%) | Male | 15 (75%) | 9 (45%) | 3.54 | 0.105 † |
| Female | 5 (25%) | 11 (55%) | |||
| Age | 62.50 ± 7.48 | 57.15 ± 13.94 | 0.141 * | ||
| Past history, n (%) | |||||
| Osteoporosis, yes | 1 (5%) | 2 (10%) | 0.48 | 0.999 † | |
| Diabetes mellitus, yes | 3 (15%) | 2 (10%) | 1.57 | 0.999 † | |
| Hypertension, yes | 6 (30%) | 1 (5%) | 7.76 | 0.091 † | |
| Hyperlipidemia, yes | 2 (10%) | 4 (20%) | 0.45 | 0.661 † | |
| BMD | 1.05 (0.2) | - | |||
| WBC (×106/uL) | 6.67 ± 2.36 | 7.62 ± 2.24 | 0.343 * | ||
| RBC (×106/uL) | 4.53 ± 0.42 | 4.54 ± 0.65 | 0.961 * | ||
| Hgb (g/dL) | 14.11 ± 1.24 | 13.46 ± 1.88 | 0.296 * | ||
| Na (mEq/L) | 140.74 ± 2.05 | 141.33 ± 2.18 | 0.487 * | ||
| K (mEq/L) | 4.28 ± 0.35 | 4.31 ± 0.46 | 0.839 * | ||
| CI (mEq/L) | 103.16 ± 1.64 | 104.00 ± 2.12 | 0.259 * | ||
| T.pro (g/dL) | 7.00 ± 0.54 | 10.39 ± 7.81 | 0.26 * | ||
| Albumin (g/dL) | 4.38 ± 0.29 | 4.53 ± 0.28 | 0.234 * | ||
| Ca (mg/dL) | 9.46 ± 0.49 | 9.57 ± 0.55 | 0.638 * | ||
| P (mg/dL) | 3.33 ± 0.41 | 3.27 ± 0.41 | 0.746 * |
| Chr | Position Type (SNP rs) | A1 | A2 | Frequency | Candidate Gene | Gene Description | OR | p-Value * | |
|---|---|---|---|---|---|---|---|---|---|
| Control (%) | RC (%) | ||||||||
| n = 20 | n = 20 | ||||||||
| 19 | 55,014,124 SNP (rs2287828) | A | G | 3 (15) | 14 (70) | LAIR2 | A membrane-bound receptor that modulates innate immune response. | 9.115942029 | 0.00054903 |
| 4 | 1,388,583 SNP (rs9328733) | A | G | 6 (30) | 13 (65) | CRIPAK | Negative regulator of PAK1. | 6 | 0.00111562 |
| 4 | 57,796,900 SNP (rs2228991) | A | G | 3 (15) | 15 (75) | REST | A transcriptional repressor that represses neuronal genes in non-neuronal tissues. | 8.222222222 | 0.00119894 |
| 19 | 9,236,698 SNP (rs111279560) | GATGGT | G | 5 (25) | 15 (75) | OR7G3 | Olfactory receptors protein of a large family of G-protein-coupled receptors (GPCR) | 5.333333333 | 0.00173965 |
| 22 | 37,584,352 SNP (rs13057424) | A | G | 6 (30) | 15 (75) | C1QTNF6 | Complement C1q tumor necrosis factor-related protein 6 | 5.210526316 | 0.00202543 |
| 4 | 57,843,295 SNP (rs3733306) | A | C | 3 (15) | 14 (70) | NOA1 | Nitric Oxide-Associated Protein 1 | 7.4 | 0.00253432 |
| 13 | 25,671,429 SNP (rs77466429) | T | G | 2 (10) | 13 (65) | PABPC3 | Poly binding protein that cytoplasmic regulatory processes of mRNA metabolism | 9.148148148 | 0.00312074 |
| 16 | 55,862,824 SNP (rs3826192) | T | C | 2 (10) | 13 (65) | CES1 | A member of the carboxylesterase large family. | 9.148148148 | 0.00312074 |
| 16 | 55,862,883 SNP (rs3826190) | A | C | 2 (10) | 13 (65) | CES1 | A member of the carboxylesterase large family. | 9.148148148 | 0.00312074 |
| 4 | 1,389,156 SNP (rs71614972) | T | C | 6 (30) | 15 (75) | CRIPAK | Negative regulator of PAK1. | 5.126984127 | 0.00334498 |
| 19 | 1,619,350 SNP (rs1052692) | T | C | 6 (30) | 15 (75) | TCF3 | A member of the E protein (class I) family of helix-loop-helix transcription factors. | 5.126984127 | 0.00334498 |
| 9 | 36,135,923 SNP (rs7041851) | C | T | 4 (20) | 12 (60) | GLIPR2 | GLI pathogenesis related 2 | 6 | 0.00379856 |
| 16 | 30,021,402 SNP (rs1140239) | T | C | 4 (20) | 14 (70) | DOC2A | Double C2 domain alpha | 5.173913043 | 0.00517988 |
| 19 | 54,744,387 SNP (rs71263238) | C | T | 4 (20) | 16 (80) | LILRB3 | A member of the leukocyte immunoglobulin-like receptor (LIR) family | 4.960784314 | 0.00564698 |
| 12 | 60,173,356 SNP (rs3763980) | T | C | 5 (25) | 16 (80) | SLC16A7 | A member of the monocarboxylate transporter family. | 4.636363636 | 0.00660224 |
| 17 | 8,161,149 SNP (rs4791641) | T | A | 6 (30) | 15 (75) | PFAS | Catalyzes biosynthesis of DNA replication, transcription, and energy metabolism | 4.636363636 | 0.00660224 |
| 17 | 35,300,494 SNP (rs35033250) | G | GA | 5 (25) | 15 (75) | LHX1 | A transcription factor of a large protein family which contains the LIM domain, | 4.363636364 | 0.00690291 |
| 1 | 197,901,206 SNP (rs34018572) | CA | C | 4 (20) | 12 (60) | LHX9 | A transcription factor of a large protein family which contains the LIM domain, | 5.1 | 0.00778198 |
| 10 | 104,240,498 SNP (rs12162) | G | A | 4 (20) | 13 (65) | ACTR1A | A macromolecular complex consisting of subunit of dynactin | 5.1 | 0.00778198 |
| 16 | 20,477,004 SNP (rs59261767) | T | C | 3 (15) | 12 (60) | ACSM2A | A macromolecular complex consisting of subunit of dynactin | 5.938271605 | 0.01028714 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, H.-J.; Kim, J.-H.; Yoon, S.; Choi, J.; Koo, J.; Lee, S. Genome-Wide Association Study Identifies Genetic Variants Associated with Rotator Cuff Tear—A Pilot Study. Diagnostics 2022, 12, 2497. https://doi.org/10.3390/diagnostics12102497
An H-J, Kim J-H, Yoon S, Choi J, Koo J, Lee S. Genome-Wide Association Study Identifies Genetic Variants Associated with Rotator Cuff Tear—A Pilot Study. Diagnostics. 2022; 12(10):2497. https://doi.org/10.3390/diagnostics12102497
Chicago/Turabian StyleAn, Hyun-Ju, Jae-Hwa Kim, Siyeong Yoon, Junwon Choi, Jeongmo Koo, and Soonchul Lee. 2022. "Genome-Wide Association Study Identifies Genetic Variants Associated with Rotator Cuff Tear—A Pilot Study" Diagnostics 12, no. 10: 2497. https://doi.org/10.3390/diagnostics12102497
APA StyleAn, H.-J., Kim, J.-H., Yoon, S., Choi, J., Koo, J., & Lee, S. (2022). Genome-Wide Association Study Identifies Genetic Variants Associated with Rotator Cuff Tear—A Pilot Study. Diagnostics, 12(10), 2497. https://doi.org/10.3390/diagnostics12102497







