Combined Locked Nucleic Acid Probes and High-Resolution Melting Curve Analysis for Detection of Rifampicin-Resistant Tuberculosis in Northern Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Mycobacterium Tuberculosis Strains and DNA Samples
2.2. The Design of H526D Probe and D516V Probe
2.3. The RIF-RDp Assay
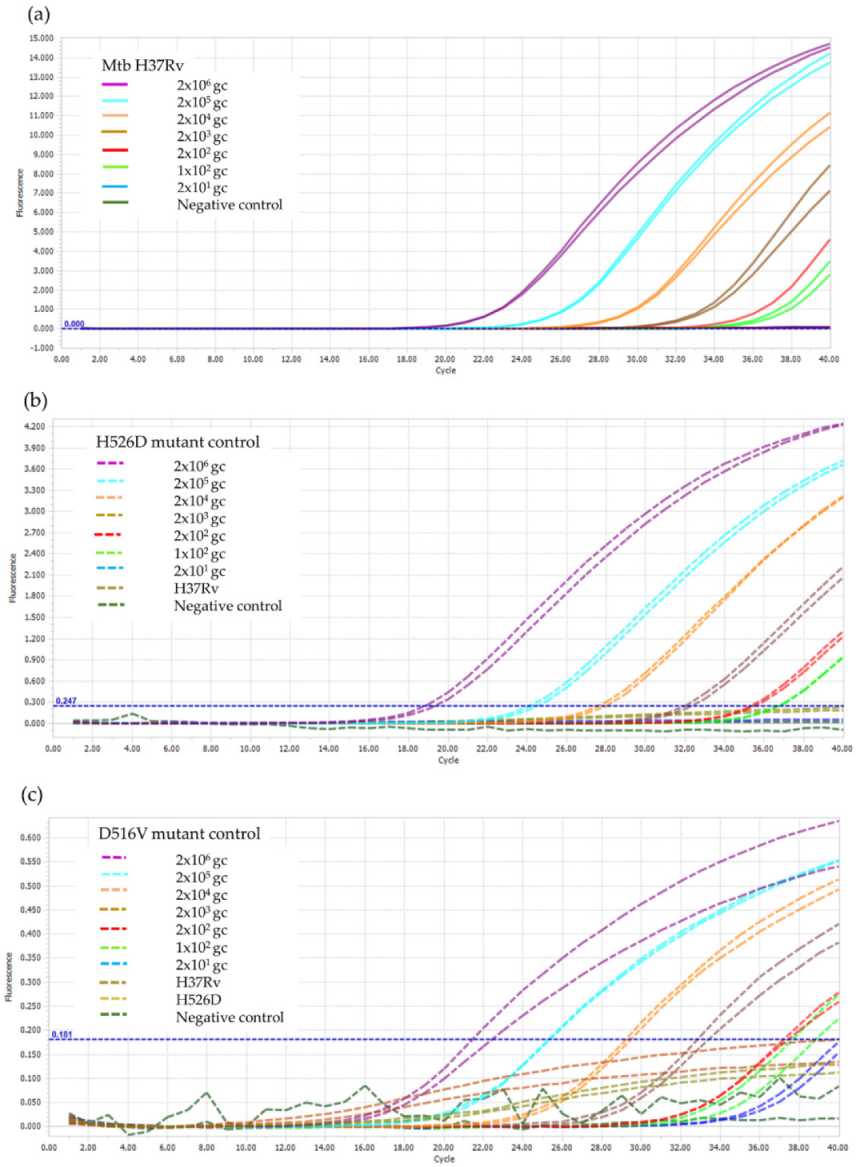
2.4. Limit of Detection (LOD) of the RIF-RDp Assay
2.5. Evaluation of the RIF-RDp Assay
2.5.1. Drug Susceptibility Testing (DST)
2.5.2. DNA Sequencing
2.5.3. Multiplex Real-Time PCR Commercial Kit
2.6. Statistical Analysis
3. Results
3.1. Optimization and the Validation of the RIF-RDp Assay
3.2. LOD of the RIF-RDp Assay
3.3. Evaluation of the RIF-RDp Assay and ROC Curve Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report. 2020. Available online: https://www.who.int/publications/i/item/9789240013131 (accessed on 30 August 2022).
- Solo, E.S.; Suzuki, Y.; Kaile, T.; Bwalya, P.; Lungu, P.; Chizimu, J.Y.; Shah, Y.; Nakajima, C. Characterization of Mycobacterium tuberculosis genotypes and their correlation to multidrug resistance in Lusaka, Zambia. Int. J. Infect. Dis. 2021, 102, 489–496. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Rapid Communication: Key Changes to Treatment of Multidrug-and Rifampicin-Resistant Tuberculosis (MDR/RR-TB); World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Woodley, C.L.; Kilburn, J.O.; David, H.L.; Silcox, V.A. Susceptibility of mycobacteria to rifampin. Antimicrob. Agents Chemother. 1972, 2, 245–249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blanchard, J.S. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu. Rev. Biochem. 1996, 65, 215–239. [Google Scholar] [CrossRef] [PubMed]
- Zaw, M.T.; Emran, N.A.; Lin, Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in Mycobacterium tuberculosis. J. Infect. Public. Health 2018, 11, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Hillemann, D.; Weizenegger, M.; Kubica, T.; Richter, E.; Niemann, S. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 2005, 43, 3699–3703. [Google Scholar] [CrossRef] [PubMed]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275. [Google Scholar] [CrossRef]
- Tomasicchio, M.; Theron, G.; Pietersen, E.; Streicher, E.; Stanley-Josephs, D.; van Helden, P.; Warren, R.; Dheda, K. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Sci. Rep. 2016, 6, 17850. [Google Scholar] [CrossRef]
- Bunsow, E.; Ruiz-Serrano, M.J.; López Roa, P.; Kestler, M.; Viedma, D.G.; Bouza, E. Evaluation of GeneXpert MTB/RIF for the detection of Mycobacterium tuberculosis and resistance to rifampin in clinical specimens. J. Infect. 2014, 68, 338–343. [Google Scholar] [CrossRef]
- World Health Organization. The Use of Next-Generation Sequencing Technologies for the Detection of Mutations Associated with Drug Resistance in Mycobacterium Tuberculosis Complex: Technical Guide; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Phelan, J.; O’Sullivan, D.M.; Machado, D.; Ramos, J.; Whale, A.S.; O’Grady, J.; Dheda, K.; Campino, S.; McNerney, R.; Viveiros, M.; et al. The variability and reproducibility of whole genome sequencing technology for detecting resistance to anti-tuberculous drugs. Genome Med. 2016, 8, 132. [Google Scholar] [CrossRef]
- Pietzka, A.T.; Indra, A.; Stöger, A.; Zeinzinger, J.; Konrad, M.; Hasenberger, P.; Allerberger, F.; Ruppitsch, W. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J. Antimicrob. Chemother. 2009, 63, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Dong, H.; Tan, Y.; Deng, Y.; Cai, X.; Jing, H.; Xia, H.; Li, Q.; Ou, X.; Su, B.; et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci. Rep. 2016, 6, 25330. [Google Scholar] [CrossRef] [PubMed]
- Anthwal, D.; Gupta, R.K.; Bhalla, M.; Bhatnagar, S.; Tyagi, J.S.; Haldar, S. Direct detection of rifampin and isoniazid resistance in sputum samples from tuberculosis patients by high-resolution melt curve analysis. J. Clin. Microbiol. 2017, 55, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.E.; Lee, S.M.; Yi, J.; Hwang, S.H.; Kim, H.H.; Lee, E.Y.; Cho, E.H.; Kim, J.H.; Kim, H.J.; Chang, C.L. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 2010, 48, 3893–3898. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Sethi, S.; Mewara, A.; Dhatwalia, S.K.; Gupta, D.; Sharma, M. Rapid detection of rifampicin, isoniazid and streptomycin resistance in Mycobacterium tuberculosis clinical isolates by high-resolution melting curve analysis. J. Appl. Microbiol. 2012, 113, 856–862. [Google Scholar] [CrossRef]
- Malhotra, B.; Goyal, S.; Bhargava, S.; Reddy, P.V.; Chauhan, A.; Tiwari, J. Rapid detection of rifampicin resistance in Mycobacterium tuberculosis by high-resolution melting curve analysis. Int. J. Tuberc. Lung Dis. 2015, 19, 1536–1541. [Google Scholar] [CrossRef]
- Anukool, U.; Phunpae, P.; Tharinjaroen, C.S.; Butr-Indr, B.; Saikaew, S.; Netirat, N.; Intorasoot, S.; Suthachai, V.; Tragoolpua, K.; Chaiprasert, A. Genotypic Distribution and a potential diagnostic assay of multidrug-resistant tuberculosis in northern Thailand. Infect. Drug Resist. 2020, 13, 3375–3382. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, G.; Sun, C.; Li, C.; Wang, X.; Liu, H.; Zhang, P.; Zhao, X.; Wang, X.; Jiang, Y.; et al. Locked nucleic acid probe-based real-time PCR assay for the rapid detection of rifampin-resistant Mycobacterium tuberculosis. PLoS ONE 2015, 10, e0143444. [Google Scholar] [CrossRef]
- Ramirez, M.V.; Cowart, K.C.; Campbell, P.J.; Morlock, G.P.; Sikes, D.; Winchell, J.M.; Posey, J.E. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 2010, 48, 4003–4009. [Google Scholar] [CrossRef]
- Prammananan, T.; Cheunoy, W.; Taechamahapun, D.; Yorsangsukkamol, J.; Phunpruch, S.; Phdarat, P.; Leechawengwong, M.; Chaiprasert, A. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin. Microbiol. Infect. 2008, 14, 446–453. [Google Scholar] [CrossRef][Green Version]
- Staroscik, A. Calculator for determining the number of copies of a template. URI Genom. Seq. Cent. 2004, 19, 2012. [Google Scholar]
- Woods, G.L.; Brown-Elliott, B.A.; Conville, P.S.; Desmond, E.P.; Hall, G.S.; Lin, G.; Pfyffer, G.E.; Ridderhof, J.C.; Siddiqi, S.H.; Wallace, R.J., Jr.; et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- XLSTAT. Data Analysis and Statistical Solution for Microsoft Excel; Addinsof Inc.: New York, NY, USA, 2017. [Google Scholar]
- O’Sullivan, D.M.; McHugh, T.D.; Gillespie, S.H. Analysis of rpoB and pncA mutations in the published literature: An insight into the role of oxidative stress in Mycobacterium tuberculosis evolution? J. Antimicrob. Chemother. 2005, 55, 674–679. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Almeida da Silva, P.E.; Palomino, J.C. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: Classical and new drugs. J. Antimicrob. Chemother. 2011, 66, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Louw, G.E.; Warren, R.M.; Gey van Pittius, N.C.; McEvoy, C.R.; Van Helden, P.D.; Victor, T.C. A balancing act: Efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 2009, 53, 3181–3189. [Google Scholar] [CrossRef]
- Draker, K.A.; Boehr, D.D.; Elowe, N.H.; Noga, T.J.; Wright, G.D. Functional annotation of putative aminoglycoside antibiotic modifying proteins in Mycobacterium tuberculosis H37Rv. J. Antibiot. 2003, 56, 135–142. [Google Scholar] [CrossRef]
- Jenkins, C.; Claxton, A.P.; Shorten, R.J.; McHugh, T.D.; Gillespie, S.H. Rifampicin resistance in tuberculosis outbreak, London, England. Emerg. Infect. Dis. 2005, 11, 931. [Google Scholar] [CrossRef]
- Prim, R.I.; Schörner, M.A.; Senna, S.G.; Nogueira, C.L.; Figueiredo, A.C.; Oliveira, J.G.; Rovaris, D.B.; Bazzo, M.L. Molecular profiling of drug resistant isolates of Mycobacterium tuberculosis in the state of Santa Catarina, southern Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 618–623. [Google Scholar] [CrossRef]
- Sinha, P.; Srivastava, G.N.; Tripathi, R.; Mishra, M.N.; Anupurba, S. Detection of mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains inhibiting wild type probe hybridization in the MTBDR plus assay by DNA sequencing directly from clinical specimens. BMC Microbiol. 2020, 20, 284. [Google Scholar] [CrossRef]
- Valim, A.R.; Rossetti, M.L.; Ribeiro, M.O.; Zaha, A. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J. Clin. Microbiol. 2000, 38, 3119–3122. [Google Scholar] [CrossRef]




| Sample Code | Mutation Profile | SNP Type | Source |
|---|---|---|---|
| Rifampicin-resistant M. tuberculosis (n = 14) | |||
| CM1 | S531L (TCG→TTG) | Class-I (n = 7) | ODPC 1 * |
| CM2 | S531L (TCG→TTG) | ODPC 1 | |
| CM3 | S531L (TCG→TTG) | ODPC 1 | |
| CM4 | S531L (TCG→TTG) | ODPC 1 | |
| CM5 | H526Y (CAC→TAC) | ODPC 1 | |
| CR1 | H526C (CAC→TGC) | CPH ** | |
| CM6 | S522L (TCG→TTG) | ODPC 1 | |
| CM7 | L511P (CTG→CGG) | Class-II (n = 3) | ODPC 1 |
| CR2 | Q513P (CAA→CCA) | CPH | |
| CR3 | H526P (CAC→CCC) | CPH | |
| CM8 | H526D (CAC→GAC) | Class-III (n = 3) | ODPC 1 |
| CR4 | H526D (CAC→GAC) | CPH | |
| CR5 | H526D (CAC→GAC) | CPH | |
| CR6 | D516V (GAC→GTC) | Class-IV (n = 1) | CPH |
| Rifampicin-susceptible M. tuberculosis(n = 20) | |||
| CM9-CM28 | No mutation | ODPC 1 | |
| Assays | RIF-RDp | PPV, NPV * (95% CI) | AnyplexTM II MTB/MDR | PPV, NPV * (95% CI) | |||
|---|---|---|---|---|---|---|---|
| R | S | R | S | ||||
| DST | R | 52 | 3 | 98.11% (88.16–99.73) | 52 | 3 | 100% |
| S | 1 | 54 | 94.74% (85.69–98.19) | 0 | 55 | 94.83% (85.92–98.22) | |
| Sensitivity, Specificity (95% CI) | 94.55% (84.88–98.86) | 98.18% (90.28–99.95) | 94.55% (84.88–98.86) | 100% (93.51–100) | |||
| DNA sequencing | R | 53 | 2 | 100% | 52 | 3 | 100% |
| S | 0 | 55 | 96.49% (87.58–99.08) | 0 | 55 | 94.83% (85.92–98.22) | |
| Sensitivity, Specificity (95% CI) | 96.36% (87.47–99.56) | 100% (93.51–100) | 94.55% (84.88–98.86) | 100% (93.51–100) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thant, Y.M.; Saikaew, S.; Tharinjaroen, C.S.; Phunpae, P.; Pongsararuk, R.; Preechasuth, K.; Butr-Indr, B.; Intorasoot, S.; Tragoolpua, K.; Chaiprasert, A.; et al. Combined Locked Nucleic Acid Probes and High-Resolution Melting Curve Analysis for Detection of Rifampicin-Resistant Tuberculosis in Northern Thailand. Diagnostics 2022, 12, 2307. https://doi.org/10.3390/diagnostics12102307
Thant YM, Saikaew S, Tharinjaroen CS, Phunpae P, Pongsararuk R, Preechasuth K, Butr-Indr B, Intorasoot S, Tragoolpua K, Chaiprasert A, et al. Combined Locked Nucleic Acid Probes and High-Resolution Melting Curve Analysis for Detection of Rifampicin-Resistant Tuberculosis in Northern Thailand. Diagnostics. 2022; 12(10):2307. https://doi.org/10.3390/diagnostics12102307
Chicago/Turabian StyleThant, Yee Mon, Sukanya Saikaew, Chayada Sitthidet Tharinjaroen, Ponrut Phunpae, Rodjana Pongsararuk, Kanya Preechasuth, Bordin Butr-Indr, Sorasak Intorasoot, Khajornsak Tragoolpua, Angkana Chaiprasert, and et al. 2022. "Combined Locked Nucleic Acid Probes and High-Resolution Melting Curve Analysis for Detection of Rifampicin-Resistant Tuberculosis in Northern Thailand" Diagnostics 12, no. 10: 2307. https://doi.org/10.3390/diagnostics12102307
APA StyleThant, Y. M., Saikaew, S., Tharinjaroen, C. S., Phunpae, P., Pongsararuk, R., Preechasuth, K., Butr-Indr, B., Intorasoot, S., Tragoolpua, K., Chaiprasert, A., & Wattananandkul, U. (2022). Combined Locked Nucleic Acid Probes and High-Resolution Melting Curve Analysis for Detection of Rifampicin-Resistant Tuberculosis in Northern Thailand. Diagnostics, 12(10), 2307. https://doi.org/10.3390/diagnostics12102307








