A Genome-Wide Association Study on Liver Stiffness Changes during Hepatitis C Virus Infection Cure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design and Study Population
2.2. Endpoint and Other Definitions
2.3. Genome-Wide Genotyping and Quality Controls
2.4. Genome-Wide Association Analysis
2.5. Gene-Based Association Study and Enrichment Analyses
2.6. Additional Statistical Analyses
3. Results
3.1. Study Population
3.2. Changes in Liver Stiffness from Baseline to SVR
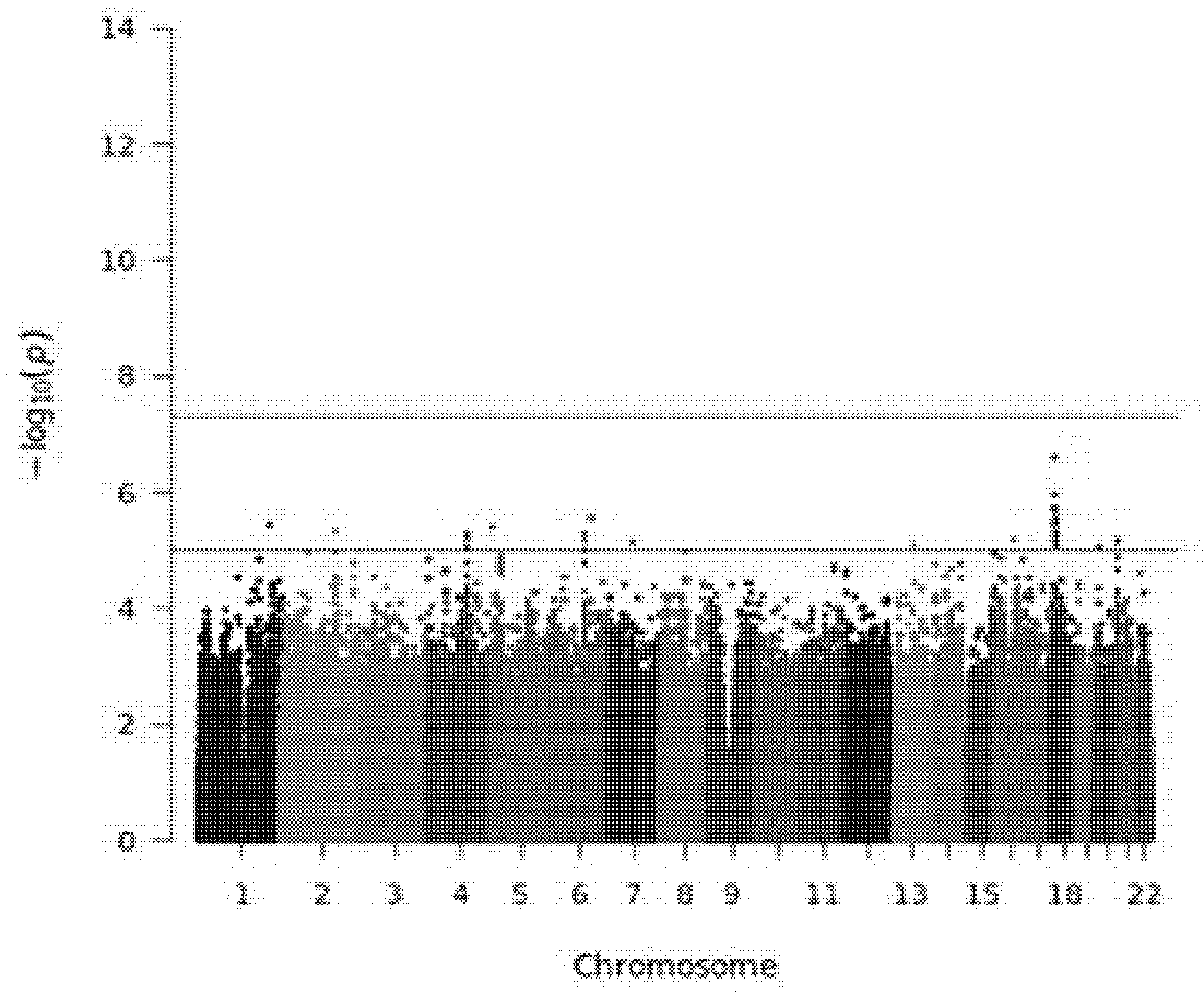
3.3. Genome-Wide Association Study
3.4. Gene-Based Association and Enrichment Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Backus, L.I.; Belperio, P.S.; Shahoumian, T.A.; Mole, L.A. Direct-acting antiviral sustained virologic response: Impact on mortality in patients without advanced liver disease. Hepatology 2018, 68, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Carrat, F.; Fontaine, H.; Dorival, C.; Simony, M.; Diallo, A.; Hezode, C.; De Ledinghen, V.; Larrey, D.; Haour, G.; Bronowicki, J.P.; et al. Clinical outcomes in patients with chronic hepatitis C after direct-acting antiviral treatment: A prospective cohort study. Lancet 2019, 393, 1453–1464. [Google Scholar] [CrossRef]
- Merchante, N.; Rivero-Juarez, A.; Tellez, F.; Merino, D.; Rios-Villegas, M.J.; Villalobos, M.; Omar, M.; Rincon, P.; Rivero, A.; Perez-Perez, M.; et al. Sustained virological response to direct-acting antiviral regimens reduces the risk of hepatocellular carcinoma in HIV/HCV-coinfected patients with cirrhosis. J. Antimicrob. Chemother. 2018, 73, 2435–2443. [Google Scholar] [CrossRef] [Green Version]
- Nahon, P.; Bourcier, V.; Layese, R.; Audureau, E.; Cagnot, C.; Marcellin, P.; Guyader, D.; Fontaine, H.; Larrey, D.; De Ledinghen, V.; et al. Eradication of Hepatitis C Virus Infection in Patients With Cirrhosis Reduces Risk of Liver and Non-Liver Complications. Gastroenterology 2017, 152, 142–156.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanwal, F.; Kramer, J.R.; Asch, S.M.; Cao, Y.; Li, L.; El-Serag, H.B. Long-Term Risk of Hepatocellular Carcinoma in HCV Patients Treated With Direct Acting Antiviral Agents. Hepatology 2020, 71, 44–55. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, A.J.; Feld, J.J.; Hofer, H.; Almasio, P.L.; Calvaruso, V.; Fernandez-Rodriguez, C.M.; Aleman, S.; Ganne-Carrie, N.; D’Ambrosio, R.; Pol, S.; et al. Risk of cirrhosis-related complications in patients with advanced fibrosis following hepatitis C virus eradication. J. Hepatol. 2017, 66, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [Green Version]
- Pawlotsky, J.; Negro, F.; Aghemo, A.; Berenguer, M.O.; Dalgard, M.; Dusheiko, G.; Marra, F.; Puoti, M.; Wedemeyer, H. EASL recommendations on treatment of hepatitis C: Final update of the series. J. Hepatol. 2020, 73, 1170–1218. [Google Scholar] [CrossRef]
- Bloom, S.; Kemp, W.; Nicoll, A.; Roberts, S.K.; Gow, P.; Dev, A.; Bell, S.; Sood, S.; Kronborg, I.; Knight, V.; et al. Liver stiffness measurement in the primary care setting detects high rates of advanced fibrosis and predicts liver-related events in hepatitis C. J. Hepatol. 2018, 69, 575–583. [Google Scholar] [CrossRef]
- Macias, J.; Marquez, M.; Tellez, F.; Merino, D.; Jimenez-Aguilar, P.; Lopez-Cortes, L.F.; Ortega, E.; von Wichmann, M.A.; Rivero, A.; Mancebo, M.; et al. Risk of liver decompensation among HIV/hepatitis C virus-coinfected individuals with advanced fibrosis: Implications for the timing of therapy. Clin. Infect. Dis. 2013, 57, 1401–1408. [Google Scholar] [CrossRef] [Green Version]
- Merchante, N.; Rivero-Juarez, A.; Tellez, F.; Merino, D.; Jose Rios-Villegas, M.; Marquez-Solero, M.; Omar, M.; Macias, J.; Camacho, A.; Perez-Perez, M.; et al. Liver stiffness predicts clinical outcome in human immunodeficiency virus/hepatitis C virus-coinfected patients with compensated liver cirrhosis. Hepatology 2012, 56, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Lledo, G.M.; Carrasco, I.; Benitez-Gutierrez, L.M.; Arias, A.; Royuela, A.; Requena, S.; Cuervas-Mons, V.; de Mendoza, C. Regression of liver fibrosis after curing chronic hepatitis C with oral antivirals in patients with and without HIV coinfection. Aids 2018, 32, 2347–2352. [Google Scholar] [CrossRef] [PubMed]
- Malin, J.J.; Boesecke, C.; Schwarze-Zander, C.; Wasmuth, J.C.; Schlabe, S.; Trebicka, J.; Spengler, U.; Llibre, J.M.; Jou, T.; Vasylyev, M.; et al. Liver stiffness regression after successful Hepatitis C treatment is independent of HIV coinfection. HIV Med. 2019, 20, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Corma-Gomez, A.; Macias, J.; Tellez, F.; Freyre-Carrillo, C.; Morano, L.; Rivero-Juarez, A.; Rios, M.J.; Alados, J.C.; Vera-Mendez, F.J.; Merchante, N.; et al. Liver Stiffness at the Time of Sustained Virological Response Predicts the Clinical Outcome in People Living With Human Immunodeficiency Virus and Hepatitis C Virus With Advanced Fibrosis Treated With Direct-acting Antivirals. Clin. Infect. Dis. 2020, 71, 2354–2362. [Google Scholar] [CrossRef]
- Corma-Gomez, A.; Morano, L.; Tellez, F.; Rivero-Juarez, A.; Real, L.M.; Alados, J.C.; Rios-Villegas, M.J.; Vera-Mendez, F.J.; Munoz, R.P.; Geijo, P.; et al. HIV infection does not increase the risk of liver complications in hepatitis C virus-infected patient with advanced fibrosis, after sustained virological response with direct-acting antivirals. Aids 2019, 33, 1167–1174. [Google Scholar] [CrossRef]
- Castera, L.; Forns, X.; Alberti, A. Non-invasive evaluation of liver fibrosis using transient elastography. J. Hepatol. 2008, 48, 835–847. [Google Scholar] [CrossRef]
- Real, L.M.; Fernandez-Fuertes, M.; Saez, M.E.; Rivero-Juarez, A.; Frias, M.; Tellez, F.; Santos, J.; Merino, D.; Moreno-Grau, S.; Gomez-Salgado, J.; et al. A genome-wide association study on low susceptibility to hepatitis C virus infection (GEHEP012 study). Liver Int. 2019, 39, 1918–1926. [Google Scholar] [CrossRef]
- Barsh, G.S.; Copenhaver, G.P.; Gibson, G.; Williams, S.M. Guidelines for genome-wide association studies. PLoS Genet. 2012, 8, e1002812. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.O. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, Y.; Shi, H.; Feng, Y.; Zhang, Z.; Zhang, L. Proteomic profiling of hepatic stellate cells in alcohol liver fibrosis reveals proteins involved in collagen production. Alcohol 2020, 86, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Cheng, X.; Sifers, R.N. Golgi-situated endoplasmic reticulum alpha-1, 2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with gamma-COP. Mol. Biol. Cell 2013, 24, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.C.; Hsiao, Y.C.; Yang, W.Y.; Tsai, S.L.; Lin, H.K.; Liao, C.Y.; Lu, J.W.; Chou, Y.T.; Wang, H.D.; Yuh, C.H. Up-regulation of golgi alpha-mannosidase IA and down-regulation of golgi alpha-mannosidase IC activates unfolded protein response during hepatocarcinogenesis. Hepatol. Commun. 2017, 1, 230–247. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Ding, J.; Wang, M.; Zhang, J.; Zhu, X.; Guan, W. Kupffer-derived matrix metalloproteinase-9 contributes to liver fibrosis resolution. Int. J. Biol. Sci. 2018, 14, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.R.; Karchner, S.I.; Allan, L.L.; Pollenz, R.S.; Tanguay, R.L.; Jenny, M.J.; Sherr, D.H.; Hahn, M.E. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: Role of DNA binding and competition for AHR nuclear translocator. Mol. Pharm. 2008, 73, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Beischlag, T.V.; Luis Morales, J.; Hollingshead, B.D.; Perdew, G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 207–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Tung, H.C.; Li, S.; Niu, Y.; Garbacz, W.G.; Lu, P.; Bi, Y.; Li, Y.; He, J.; Xu, M.; et al. Aryl Hydrocarbon Receptor Signaling Prevents Activation of Hepatic Stellate Cells and Liver Fibrogenesis in Mice. Gastroenterology 2019, 157, 793–806.e14. [Google Scholar] [CrossRef]
- Speliotes, E.K.; Yerges-Armstrong, L.M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D.; Gudnason, V.; Eiriksdottir, G.; Garcia, M.E.; Launer, L.J.; et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Patin, E.; Kutalik, Z.; Guergnon, J.; Bibert, S.; Nalpas, B.; Jouanguy, E.; Munteanu, M.; Bousquet, L.; Argiro, L.; Halfon, P.; et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology 2012, 143, 1244–1252.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulveling, D.; Le Clerc, S.; Cobat, A.; Labib, T.; Noirel, J.; Laville, V.; Coulonges, C.; Carpentier, W.; Nalpas, B.; Heim, M.H.; et al. A new 3p25 locus is associated with liver fibrosis progression in human immunodeficiency virus/hepatitis C virus-coinfected patients. Hepatology 2016, 64, 1462–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamdane, N.; Juhling, F.; Crouchet, E.; El Saghire, H.; Thumann, C.; Oudot, M.A.; Bandiera, S.; Saviano, A.; Ponsolles, C.; Roca Suarez, A.A.; et al. HCV-Induced Epigenetic Changes Associated with Liver Cancer Risk Persist After Sustained Virologic Response. Gastroenterology 2019, 156, 2313–2329.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, S.; Kaspi, A.; Domovitz, T.; Davidovich, A.; Lavi-Itzkovitz, A.; Meirson, T.; Alison Holmes, J.; Dai, C.Y.; Huang, C.F.; Chung, R.T.; et al. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet. 2019, 15, e1008181. [Google Scholar] [CrossRef]
- Corma-Gómez, A.; Macías, J.; Lacalle-Remigio, J.R.; Téllez, F.; Morano, L.; Rivero, A.; Serrano, M.; Ríos, M.J.; Vera-Méndez, F.J.; Alados, J.C.; et al. HIV infection is associated with lower risk of hepatocellular carcinoma after sustained virological response to direct-acting antivirals in hepatitis C infected-patients with advanced fibrosis. Clin. Infect. Dis. 2020, in press. [Google Scholar] [CrossRef]
- Rinaldi, L.; Perrella, A.; Guarino, M.; De Luca, M.; Piai, G.; Coppola, N.; Pafundi, P.C.; Ciardiello, F.; Fasano, M.; Martinelli, E.; et al. Incidence and risk factors of early HCC occurrence in HCV patients treated with direct acting antivirals: A prospective multicentre study. J. Transl. Med. 2019, 17, 292–301. [Google Scholar] [CrossRef]
| Variables | GWAS Population (n = 242) | Gehep-011 Entire Cohort (n = 1035) | p |
|---|---|---|---|
| Age, years † | 50 (46–53) | 51 (47–55) | 0.019 |
| Males, n (%) | 202 (83.5) | 844 (81.5) | 0.484 |
| HIV infection, n (%) | 174 (71.9) | 667 (64.4) | 0.028 |
| PWID, n (%) | 181 (74.8) | 697 (67.3) | 0.025 |
| HCV genotype 3, n (%) | 41 (16.9) | 178 (17.2) | 0.919 |
| HCV viral load † ‡ | 163 (68–398) | 177 (59–415) | 0.854 |
| IFN-free treatment, n (%) | 208 (86) | 973 (94) | <0.001 |
| Baseline liver stiffness (KPa) † | 116.8 (11.8–27.7) | 16.8 (11.8–26.7) | 0.487 |
| CHR | SNP | BP | A1 | MAF | BETA (95%CI) | p | Gene |
|---|---|---|---|---|---|---|---|
| 18 | rs12606769 | 9012996 | C | 0.207 | −13.27 (−18.16–−8.386) | 2.48 × 10−7 | NDUFV2 † |
| 18 | rs9959475 | 9009311 | A | 0.234 | −12.14 (−16.89–−7.387) | 1.10 × 10−6 | NDUFV2 † |
| 18 | rs1553736 | 9012842 | C | 0.215 | −12.21 (−17.09–−7.333) | 1.80 × 10−6 | NDUFV2 † |
| 18 | rs56232039 | 9013366 | A | 0.212 | −12.23 (−17.14–−7.315) | 2.05 × 10−6 | NDUFV2 † |
| 6 | rs2649545 | 119771886 | A | 0.447 | −10.98 (−15.45–−6.508) | 2.77 × 10−6 | MAN1A1 † |
| 18 | rs67749125 | 11936826 | T | 0.048 | −25.16 (−35.44–−14.89) | 2.91 × 10−6 | MPPE1 † |
| 18 | rs8095587 | 11936664 | A | 0.048 | −25.16 (−35.44–−14.89) | 2.91 × 10−6 | MPPE1 † |
| 18 | rs12326768 | 11925795 | G | 0.052 | −24.06 (−33.96–−14.17) | 3.37 × 10−6 | MPPE1 † |
| 18 | rs72942777 | 9004175 | T | 0.268 | −10.78 (−15.22–−6.344) | 3.45 × 10−6 | NDUFV2 † |
| 1 | rs188739258 | 208743339 | T | 0.013 | −42.14 (−59.53–−24.75) | 3.60 × 10−6 | |
| 5 | rs62330020 | 311880 | T | 0.047 | −22.65 (−32.03–−13.26) | 3.94 × 10−6 | AHRR |
| 2 | rs6437198 | 159613196 | C | 0.277 | −11.38 (−16.13–−6.625) | 4.70 × 10−6 | APL1 † |
| 18 | rs12955366 | 11922625 | C | 0.056 | −23.79 (−33.75–−13.83) | 4.90 × 10−6 | MPPE1 † |
| 18 | rs9951113 | 11927429 | C | 0.050 | −24.12 (−34.22–−14.01) | 4.95 × 10−6 | MPPE1 † |
| 6 | rs9402699 | 99800122 | A | 0.277 | −11.49 (−16.31–−6.662) | 5.19 × 10−6 | FAXC † |
| 4 | rs8180156 | 115907555 | C | 0.110 | 16.62 (9.637–23.6) | 5.22 × 10−6 | NDST4 |
| 18 | rs9962961 | 11936268 | C | 0.050 | −24.11 (−34.24–−13.98) | 5.25 × 10−6 | MPPE1 † |
| 18 | rs112570549 | 11920486 | T | 0.051 | −23.43 (−33.33–−13.53) | 5.95 × 10−6 | MPPE1 † |
| 4 | rs17623036 | 115902626 | C | 0.108 | 16.61 (9.57–23.66) | 6.32 × 10−6 | NDST4 † |
| 16 | rs74918996 | 53055101 | C | 0.022 | −33.49 (−47.71–−19.27) | 6.55 × 10−6 | CHD9 † |
| 6 | rs6924993 | 99798835 | G | 0.281 | −11.37 (−16.21–−6.538) | 6.69 × 10−6 | COQ3 † |
| 20 | rs6122460 | 62100105 | A | 0.096 | −15.91 (−22.68–−9.141) | 6.83 × 10−6 | EEF1A2 † |
| 18 | rs11081454 | 9005806 | A | 0.257 | −10.86 (−15.49–−6.238) | 6.96 × 10−6 | NDUFV2 † |
| 20 | rs310602 | 62109170 | A | 0.101 | −15.78 (−22.51–−9.059) | 7.07 × 10−6 | EEF1A2 † |
| 7 | rs71537604 | 73067749 | T | 0.071 | −18.85 (−26.9–−10.81) | 7.30 × 10−6 | VPS37D † |
| 18 | rs56786794 | 11922278 | A | 0.057 | −22.95 (−32.76–−13.14) | 7.53 × 10−6 | MPPE1 † |
| 13 | rs61971490 | 73064151 | T | 0.045 | −24.11 (−34.45–−13.77) | 8.10 × 10−6 | |
| 18 | rs12960421 | 11923803 | G | 0.058 | −22.89 (−32.72–−13.06) | 8.23 × 10−6 | MPPE1 † |
| 20 | rs4815993 | 7263756 | A | 0.348 | 10.09 (5.749–14.43) | 8.62 × 10−6 | |
| 4 | rs114558514 | 115943168 | T | 0.108 | 16.51 (9.389–23.63) | 8.99 × 10−6 | NDST4 † |
| 4 | rs74700222 | 115938513 | G | 0.108 | 16.51 (9.389–23.63) | 8.99 × 10−6 | NDST4 |
| 4 | rs76144590 | 115936946 | G | 0.108 | 16.51 (9.389–23.63) | 8.99 × 10−6 | NDST4 |
| 4 | rs77275720 | 115946034 | C | 0.108 | 16.54 (9.395–23.68) | 9.26 × 10−6 | NDST4 |
| 6 | rs13201542 | 99934459 | A | 0.076 | −17.47 (−25.04–−9.894) | 9.96 × 10−6 | USP45 |
| 6 | rs28385588 | 99915661 | G | 0.076 | −17.47 (−25.04–−9.894) | 9.96 × 10−6 | USP45 |
| Process Description | GO Term | Number of Genes in the Term | FDR p-Value | Overloaded Genes |
|---|---|---|---|---|
| DNA packaging | GO:0006323 | 221 | <0.001 | NCAPH, HIST2H2BE, HIST2H4B, SMC2, HIST2H3A, H2AFY, PRM2, PRM1, PRM3, CDAN1, TNP2 |
| DNA conformation change | GO:0071103 | 325 | 0.001 | NCAPH, HIST2H2BE, HIST2H4B, SMC2, HIST2H3A, H2AFY, PRM2, PRM1, PRM3, CDAN1, TNP2, RAD23B |
| Chromosome organization | GO:0051276 | 1293 | 0.007 | H2AFY, STAG1, NCAPH, MSL2, HIST2H2AC, HIST2H2AB, HIST2H2BE, HIST2H4B, HIST2H2AA4, RAD23B, SMC2, RNF20, HIST2H3A, ATXN3, RBL2, PRM2, SETD5, PPM1D, PRM1, PRM3, CDAN1, TNP2 |
| Chromatin organization | GO:0006325 | 849 | 0.039 | H2AFY, MSL2, HIST2H2AC, HIST2H2AB, HIST2H2BE, HIST2H4B, HIST2H2AA4, RNF20, HIST2H3A, ATXN3, RBL2, SETD5, PPM1D, CDAN1, TNP2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corma-Gómez, A.; Macías, J.; Rivero, A.; Rivero-Juarez, A.; de los Santos, I.; Reus-Bañuls, S.; Morano, L.; Merino, D.; Palacios, R.; Galera, C.; et al. A Genome-Wide Association Study on Liver Stiffness Changes during Hepatitis C Virus Infection Cure. Diagnostics 2021, 11, 1501. https://doi.org/10.3390/diagnostics11081501
Corma-Gómez A, Macías J, Rivero A, Rivero-Juarez A, de los Santos I, Reus-Bañuls S, Morano L, Merino D, Palacios R, Galera C, et al. A Genome-Wide Association Study on Liver Stiffness Changes during Hepatitis C Virus Infection Cure. Diagnostics. 2021; 11(8):1501. https://doi.org/10.3390/diagnostics11081501
Chicago/Turabian StyleCorma-Gómez, Anaïs, Juan Macías, Antonio Rivero, Antonio Rivero-Juarez, Ignacio de los Santos, Sergio Reus-Bañuls, Luis Morano, Dolores Merino, Rosario Palacios, Carlos Galera, and et al. 2021. "A Genome-Wide Association Study on Liver Stiffness Changes during Hepatitis C Virus Infection Cure" Diagnostics 11, no. 8: 1501. https://doi.org/10.3390/diagnostics11081501
APA StyleCorma-Gómez, A., Macías, J., Rivero, A., Rivero-Juarez, A., de los Santos, I., Reus-Bañuls, S., Morano, L., Merino, D., Palacios, R., Galera, C., Fernández-Fuertes, M., González-Serna, A., de Rojas, I., Ruiz, A., Sáez, M. E., Real, L. M., & Pineda, J. A. (2021). A Genome-Wide Association Study on Liver Stiffness Changes during Hepatitis C Virus Infection Cure. Diagnostics, 11(8), 1501. https://doi.org/10.3390/diagnostics11081501







