Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of CTNNB1 S37C Plasmid
2.2. Study Subjects and Samples
2.3. Tissue DNA Isolation and Quantitation
2.4. Urine Collection and DNA Isolation and Fractionation
2.5. Detection of CTNNB1 Codon 32–37 Hotspot Mutations
2.6. Sanger Sequencing
3. Results
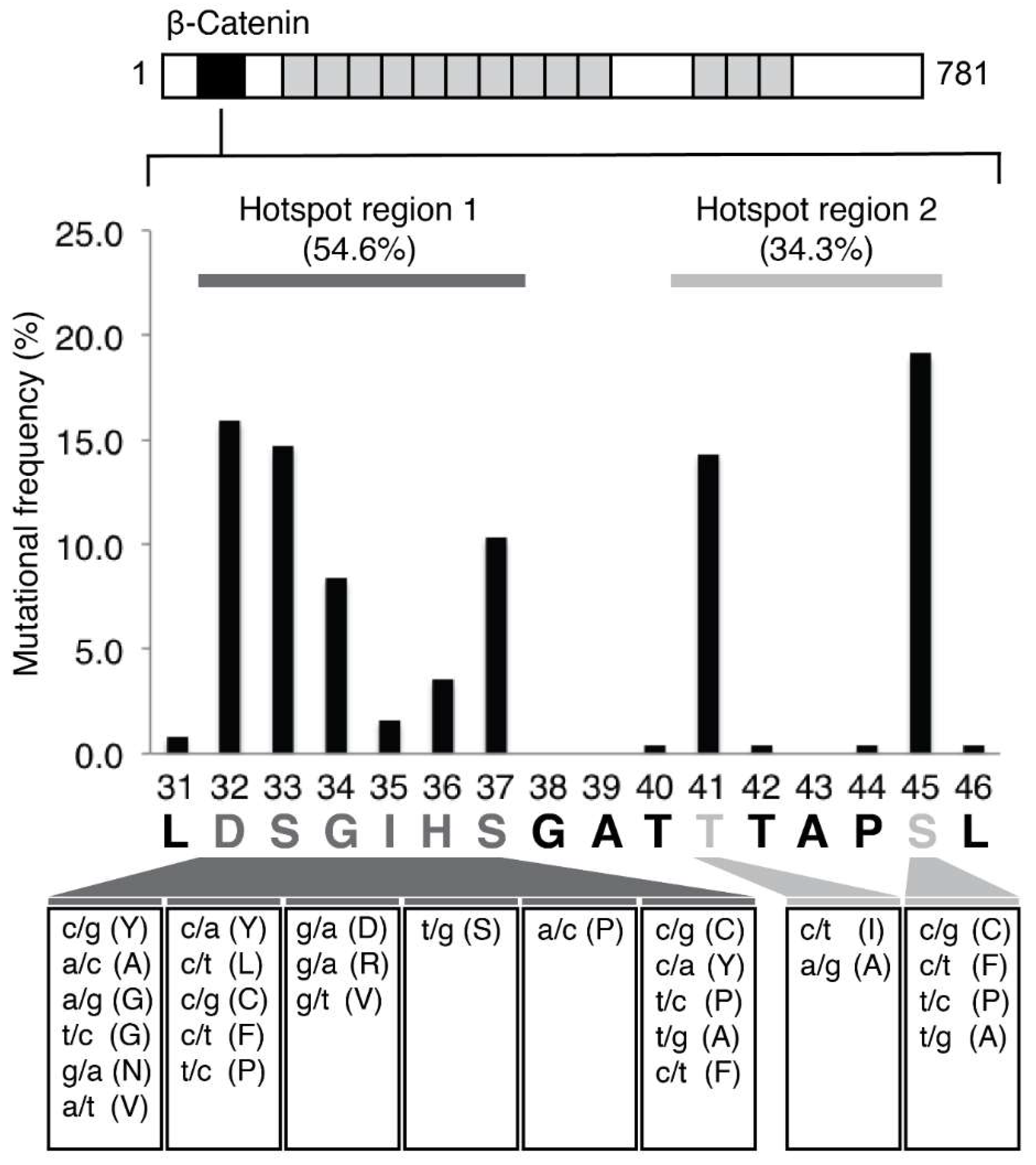
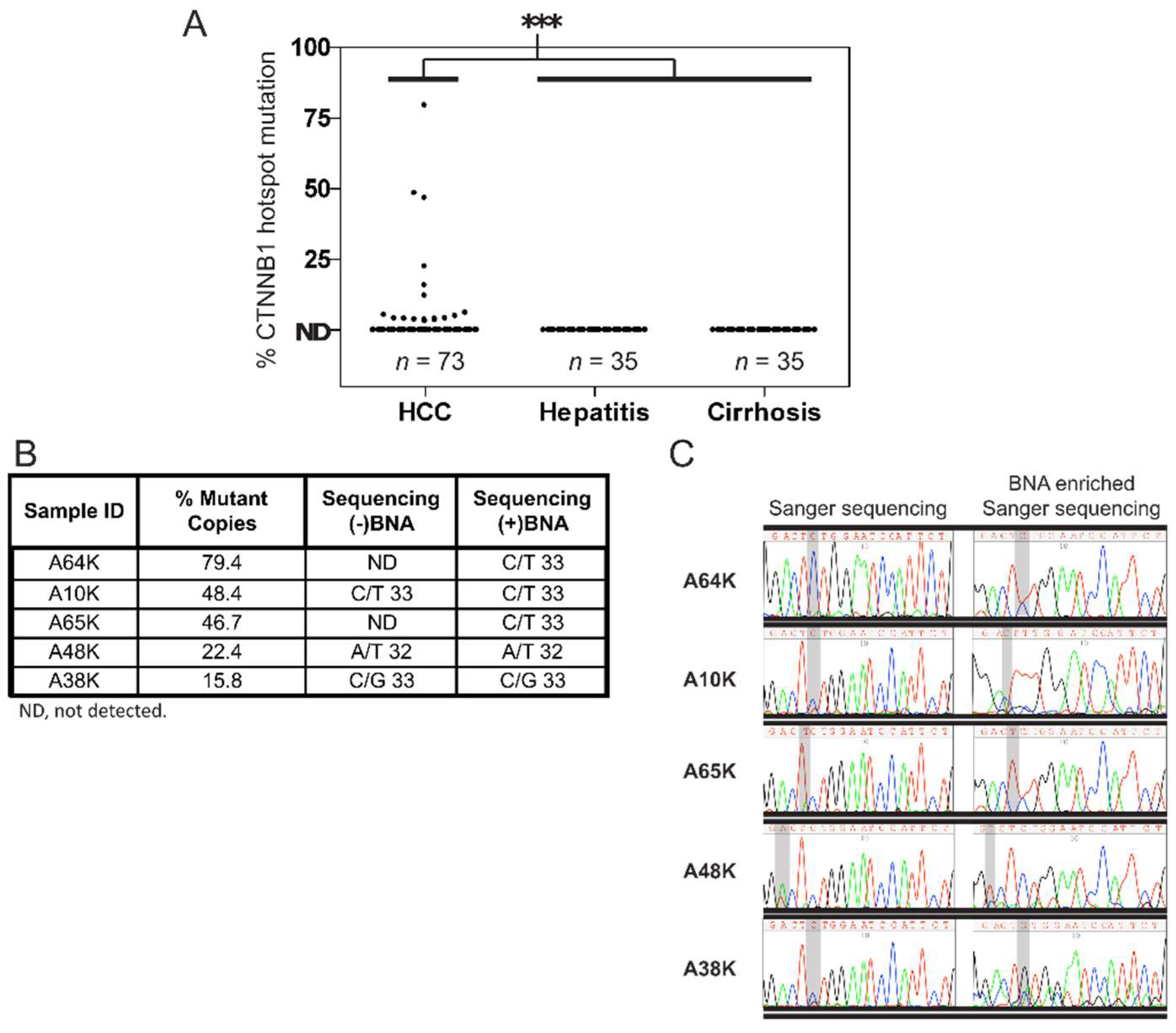
3.1. Detection of CTNNB1 Hotspot Mutations in HCC and Non-HCC Tissues
3.2. Detection of CTNNB1 Hotspot Mutations in Urine of HCC Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Kim, A.K.; Jain, S. Liquid biopsies for hepatocellular carcinoma. Transl. Res. 2018, 201, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wu, X.; Lin, C.; Zhang, X.; Ye, L.; Ren, L.; Chen, M.; Yang, M.; Li, Y.; Li, M.; et al. AKIP1 promotes early recurrence of hepatocellular carcinoma through activating the Wnt/β-catenin/CBP signaling pathway. Oncogene 2019, 38, 5516–5529. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Mu, X.D.; Song, J.R.; Zhai, P.T.; Cheng, Y.; Le, Y.; Li, Z.B. PAF enhances cancer stem cell properties via β-catenin signaling in hepatocellular carcinoma. Cell Cycle 2021, 20, 1010–1020. [Google Scholar] [CrossRef]
- Lee, J.S. The mutational landscape of hepatocellular carcinoma. Clin. Mol. Hepatol. 2015, 21, 220–229. [Google Scholar] [CrossRef] [Green Version]
- Shibata, T. Genomic landscape of hepatocarcinogenesis. J. Hum. Genet. 2021. [Google Scholar] [CrossRef]
- Teufel, M.; Seidel, H.; Köchert, K.; Meinhardt, G.; Finn, R.S.; Llovet, J.M.; Bruix, J. Biomarkers associated with response to regorafenib in patients with hepatocellular carcinoma. Gastroenterology 2019, 156, 1731–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, J.J.; Nandakumar, S.; Armenia, J.; Khalil, D.N.; Albano, M.; Ly, M.; Shia, J.; Hechtman, J.F.; Kundra, R.; El Dika, I. Prospective genotyping of hepatocellular carcinoma: Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin. Cancer Res. 2019, 25, 2116–2126. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.H.; Lin, S.Y.; Song, W.; Jain, S. DNA markers in molecular diagnostics for hepatocellular carcinoma. Expert Rev. Mol. Diagn. 2014, 14, 803–817. [Google Scholar] [CrossRef] [Green Version]
- Delgado, E.; Okabe, H.; Preziosi, M.; Russell, J.O.; Alvarado, T.F.; Oertel, M.; Nejak-Bowen, K.N.; Zhang, Y.; Monga, S.P. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J. Hepatol 2015, 62, 380–387. [Google Scholar] [CrossRef] [Green Version]
- Lachenmayer, A.; Alsinet, C.; Savic, R.; Cabellos, L.; Toffanin, S.; Hoshida, Y.; Villanueva, A.; Minguez, B.; Newell, P.; Tsai, H.W.; et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin. Cancer Res. 2012, 18, 4997–5007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashihara, E.; Takada, T.; Maekawa, T. Targeting the canonical Wnt/β-catenin pathway in hematological malignancies. Cancer Sci. 2015, 106, 665–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, R.; Mishra, D.P. Pharmacological modulation of beta-catenin and its applications in cancer therapy. J. Cell. Mol. Med. 2013, 17, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Portolani, N.; Coniglio, A.; Ghidoni, S.; Giovanelli, M.; Benetti, A.; Tiberio, G.A.; Giulini, S.M. Early and late recurrence after liver resection for hepatocellular carcinoma: Prognostic and therapeutic implications. Ann. Surg. 2006, 243, 229–235. [Google Scholar] [CrossRef]
- Shah, S.A.; Cleary, S.P.; Wei, A.C.; Yang, I.; Taylor, B.R.; Hemming, A.W.; Langer, B.; Grant, D.R.; Greig, P.D.; Gallinger, S. Recurrence after liver resection for hepatocellular carcinoma: Risk factors, treatment, and outcomes. Surgery 2007, 141, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M. Recurrence of hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 2045–2047. [Google Scholar] [CrossRef]
- Takeishi, K.; Maeda, T.; Tsujita, E.; Yamashita, Y.; Harada, N.; Itoh, S.; Harimoto, N.; Ikegami, T.; Yoshizumi, T.; Shirabe, K.; et al. Predictors of intrahepatic multiple recurrences after curative hepatectomy for hepatocellular carcinoma. Anticancer. Res. 2015, 35, 3061–3066. [Google Scholar] [PubMed]
- Shah, S.A.; Greig, P.D.; Gallinger, S.; Cattral, M.S.; Dixon, E.; Kim, R.D.; Taylor, B.R.; Grant, D.R.; Vollmer, C.M. Factors associated with early recurrence after resection for hepatocellular carcinoma and outcomes. J. Am. Coll. Surg. 2006, 202, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Song, B.P.; Jain, S.; Lin, S.Y.; Chen, Q.; Block, T.M.; Song, W.; Brenner, D.E.; Su, Y.-H. Detection of hypermethylated vimentin in urine of patients with colorectal cancer. J. Mol. Diagn. 2012, 14, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.Y.; Dhillon, V.; Jain, S.; Chang, T.-T.; Hu, C.-T.; Lin, Y.-J.; Chen, S.-H.; Chang, K.-C.; Song, W.; Yu, L. A locked nucleic acid clamp-mediated PCR assay for detection of a p53 codon 249 hotspot mutation in urine. J. Mol. Diagn. 2011, 13, 474–484. [Google Scholar] [CrossRef]
- Su, Y.H.; Song, J.; Wang, Z.; Wang, X.H.; Wang, M.; Brenner, D.E.; Block, T.M. Removal of high-molecular-weight DNA by carboxylated magnetic beads enhances the detection of mutated K-ras DNA in urine. Ann. N. Y. Acad Sci. 2008, 1137, 82–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornesello, M.L.; Buonaguro, L.; Tatangelo, F.; Botti, G.; Izzo, F.; Buonaguro, F.M. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics 2013, 102, 74–83. [Google Scholar] [CrossRef] [Green Version]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.; Lee, D.; Ma, Y. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hann, H.-W.; Jain, S.; Park, G.; Steffen, J.D.; Song, W.; Su, Y.-H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res. 2017, 3, 105–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Xie, L.; Boldbaatar, B.; Lin, S.Y.; Hamilton, J.P.; Meltzer, S.J.; Chen, S.-H.; Hu, C.-T.; Block, T.M.; Song, W.; et al. Differential methylation of the promoter and first exon of the RASSF1A gene in hepatocarcinogenesis. Hepatol. Res. 2014, 45, 1110–1123. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Lim, J.S.; Kurzrock, R. Analysis of tissue and circulating tumor DNA by next-generation sequencing of hepatocellular carcinoma: Implications for targeted therapeutics. Mol. Cancer Ther. 2018, 17, 1114–1122. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Tsigelny, I.F.; Skjevik, Å.A.; Kono, Y.; Mendler, M.; Kuo, A.; Sicklick, J.K.; Heestand, G.; Banks, K.C.; Talasaz, A. Next-generation sequencing of circulating tumor DNA reveals frequent alterations in advanced hepatocellular carcinoma. Oncol. 2018, 23, 586–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, A.; Zhang, X.; Zhou, S.-L.; Cao, Y.; Huang, X.-W.; Fan, J.; Yang, X.-R.; Zhou, J. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J. Cancer 2016, 7, 1907–1914. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Yang, H.; Xu, H.; Wang, Y.; Ge, P.; Ren, J.; Xu, W.; Lu, X.; Sang, X.; Zhong, S. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016, 7, 40481–40490. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Huang, A.; Wang, Y.-P.; Yin, Y.; Fu, P.-Y.; Zhang, X.; Zhou, J. Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma. Ann. Transl. Med. 2020, 8, 237–249. [Google Scholar] [CrossRef]
- Lin, S.Y.; Luo, Y.; Marshall, M.M.; Johnson, B.J.; Park, S.R.; Wang, Z.; Su, Y.-H. A New Method for improving extraction efficiency and purity of urine and plasma cell-free DNA. Diagnostics 2021, 11, 650. [Google Scholar] [CrossRef]
- Su, Y.-H.; Wang, M.; Brenner, D.E.; Norton, P.A.; Block, T.M. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann. N. Y. Acad. Sci. 2008, 1137, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Galarreta, M.R.; Bresnahan, E.; Molina-Sánchez, P.; Lindblad, K.E.; Maier, B.; Sia, D.; Puigvehi, M.; Miguela, V.; Casanova-Acebes, M.; Dhainaut, M. β-catenin activation promotes immune escape and resistance to anti–PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019, 9, 1124–1141. [Google Scholar] [CrossRef] [PubMed]
- Jelic, S.; Sotiropoulos, G.C.; the ESMO Guidelines Working Group. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010, 21, v59–v64. [Google Scholar] [CrossRef]
- Xue, R.; Li, R.; Guo, H.; Guo, L.; Su, Z.; Ni, X.; Qi, L.; Zhang, T.; Li, Q.; Zhang, Z.; et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2015, 150, 998–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Diagnosis | Hepatitis (n = 35) | Cirrhosis (n = 35) | HCC (n = 73) |
|---|---|---|---|
| Mean age ± SD years | 54 ± 12 | 56 ± 14 | 60 ± 12 |
| Gender (Male:Female:Unknown) | 17:18:0 | 23:12:0 | 45:20:8 |
| Etiology | |||
| HBV | 3 | 6 | 31 |
| HCV | 22 | 17 | 19 |
| HBV/HCV | 9 | 12 | 2 |
| Other | 0 | 0 | 14 |
| Unknown | 1 | 0 | 7 |
| Stage ^ | NA | NA | |
| 1 | 19 | ||
| 2 | 31 | ||
| 3 | 12 | ||
| 4 | 11 | ||
| Grade # | NA | NA | |
| 1 | 9 | ||
| 2 | 41 | ||
| 3 | 15 | ||
| Unknown | 8 | ||
| Tumor size, mean ± SD, cm | NA | NA | 5.0±3.3 |
| AFP level, ng/mL | NA | NA | |
| ≤20 | 28 | ||
| >20 | 37 | ||
| Unknown | 8 |
| Diagnosis | HCC (n = 62) |
|---|---|
| Mean age ± SD years | 59.9 ± 11.4 |
| Gender (Male:Female) | 44:18 |
| Etiology | |
| HBV | 30 |
| HCV | 15 |
| HBV/HCV | 1 |
| Other | 10 |
| Unknown | 6 |
| Stage * | |
| 1 | 18 |
| 2 | 28 |
| 3 | 12 |
| 4 | 2 |
| Unknown | 2 |
| Grade # | |
| 1 | 8 |
| 2 | 38 |
| 3 | 14 |
| Unknown | 2 |
| Tumor size, mean ± SD, cm | 5.26 ± 3.27 |
| AFP level, ng/mL | |
| ≤20 | 38 |
| >20 | 24 |
| Unknown | 0 |
| Sample ID | Serum AFP (ng/mL) | Tumor | CTNNB1 32–37 Mutation (Copies per mL of Urine) # | Urine Collection Post-Tumor Resection (Months) | Recurrence ^ | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stage * | Grade # | Size (cm) | Before Resection | After Resection | Detected | Months Post-Resection | |||
| U1 | 6.9 | 1 | G2 | 1.9 | 2–20 | ND | 10 | No | NA |
| U2 | 5.0 | 1 | G2 | 3.5 | 2–20 | 29 | 3 | Yes | 26 |
| U3 | 19.1 | 2 | G3 | 14.5 | 2–20 | 881 | 3 | Lung metastasis | 13 |
| U4 | 1.8 | 3A | G2 | 8.0 | 2–20 | 23 | 2 | Yes | 51 |
| U5 | 11.7 | 2 | G3 | 4.4 | 2–20 | 2–20 | 2 | Yes | 21 |
| U6 | 6.5 | 3A | G2 | 3.5 | 21 | ND | 2 | No | NA |
| U7 | NT | 2 | G2 | 1.5 | 24 | ND | 10 | Yes | 21 |
| U8 | 3.8 | 1 | G2 | 1.5 | 29 | ND | 2 | No | NA |
| U9 | 6101.0 | 2 | G2 | 3.0 | 39 | ND | 1 | Yes | 4 |
| U10 | 4.0 | 1 | G1 | 2.0 | 42 | ND | 3 | No | NA |
| U11 | <1.5 | 3C | G3 | 6.0 | 101 | 34 | 2 | Yes | 7 |
| U12 | NT | 1 | G2 | 4.0 | 142 | 26 | 1 | Yes | 47 |
| U13 | 4.3 | 3A | G3 | 5.0 | 498 | ND | 1 | Yes | 57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.Y.; Chang, T.-T.; Steffen, J.D.; Chen, S.; Jain, S.; Song, W.; Lin, Y.-J.; Su, Y.-H. Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients. Diagnostics 2021, 11, 1475. https://doi.org/10.3390/diagnostics11081475
Lin SY, Chang T-T, Steffen JD, Chen S, Jain S, Song W, Lin Y-J, Su Y-H. Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients. Diagnostics. 2021; 11(8):1475. https://doi.org/10.3390/diagnostics11081475
Chicago/Turabian StyleLin, Selena Y., Ting-Tsung Chang, Jamin D. Steffen, Sitong Chen, Surbhi Jain, Wei Song, Yih-Jyh Lin, and Ying-Hsiu Su. 2021. "Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients" Diagnostics 11, no. 8: 1475. https://doi.org/10.3390/diagnostics11081475
APA StyleLin, S. Y., Chang, T.-T., Steffen, J. D., Chen, S., Jain, S., Song, W., Lin, Y.-J., & Su, Y.-H. (2021). Detection of CTNNB1 Hotspot Mutations in Cell-Free DNA from the Urine of Hepatocellular Carcinoma Patients. Diagnostics, 11(8), 1475. https://doi.org/10.3390/diagnostics11081475






