Predicting Hemodynamic Failure Development in PICU Using Machine Learning Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Study Population
2.2. Machine Learning Techniques
- Generalized linear models (GLM) are frequently used to analyze binary data. We specified a complementary log-log link (clog log) given that an asymmetric link function is more appropriate when imbalanced data are analyzed [14]. We compared the GLM (clog log) model with machine learning techniques (MLTs) to see whether the ML approaches can deliver more remarkable results.
- RPART can be used for classification and regression tasks. Breiman proposed the classification and regression tree (CART), the best-known methodology for constructing decision trees (DT) [15]. The basic goal of a DT is to repeatedly decompose data into smaller subsets using a set of splitting rules until a specific stopping criterion is encountered. The key advantage of this method is that the tree structure is easily interpretable; however, this algorithm is prone to over-fitting [16].
- Random forest is a supervised ensemble learning method that builds a collection of decision trees obtained via bootstrap aggregation, resulting in a forest of trees to predict the outcome of interest. The RF model turns out to be less interpretable than DT but improves the robustness of predictions [17].
- In its simplest form, the neural network model is composed of an input layer, a hidden layer, and an output layer (three layers of neurons which are connected). The features extracted by the model represent the input layer and are used to predict an output. The nodes displayed in the input layer communicate with each node in the hidden layer, which is connected to an output layer. The purpose is to compute a weighted sum based on ‘neurons’ importance and to provide an output [18].
- A single-hidden-layer neural network (NNET) representing the simplest form of the neural network, in which there is only one layer of input nodes that send weighted inputs to a subsequent layer of receiving nodes has been considered for the computation.
- XGB is a decision-tree-based ensemble machine learning algorithm that can be used for classification or regression problems, increasing model accuracy. XGB uses a gradient boosting framework; the idea is to build trees sequentially in such a way as to ensure that the errors of the previous three are reduced; on the contrary, RF combines results at the end of the process training independently each classifier [19].
2.3. Imbalance Control and Missing Data Imputation Techniques
2.4. Missing Data Imputation
2.5. Model Training and Validation
- Fit a model on the whole data set.
- Calculate the apparent performance of this model corresponding to the error that the model demonstrates on the original dataset.
- Create 100 bootstrap resamples.
- For every bootstrap resample, fit the model on that resample, calculate the apparent performance for this model on the bootstrap resample it was trained on, and find the apparent performance on the original dataset.
- Calculate the optimism by performing the difference between the apparent performance obtained within the bootstrap resample and the apparent performance obtained on the original dataset.
- Calculate the average optimism from all of the bootstrap samples.
- The final performance is achieved by computing the difference between the original data apparent performance and the optimism measure [26].
2.6. Statistical Analysis
3. Results
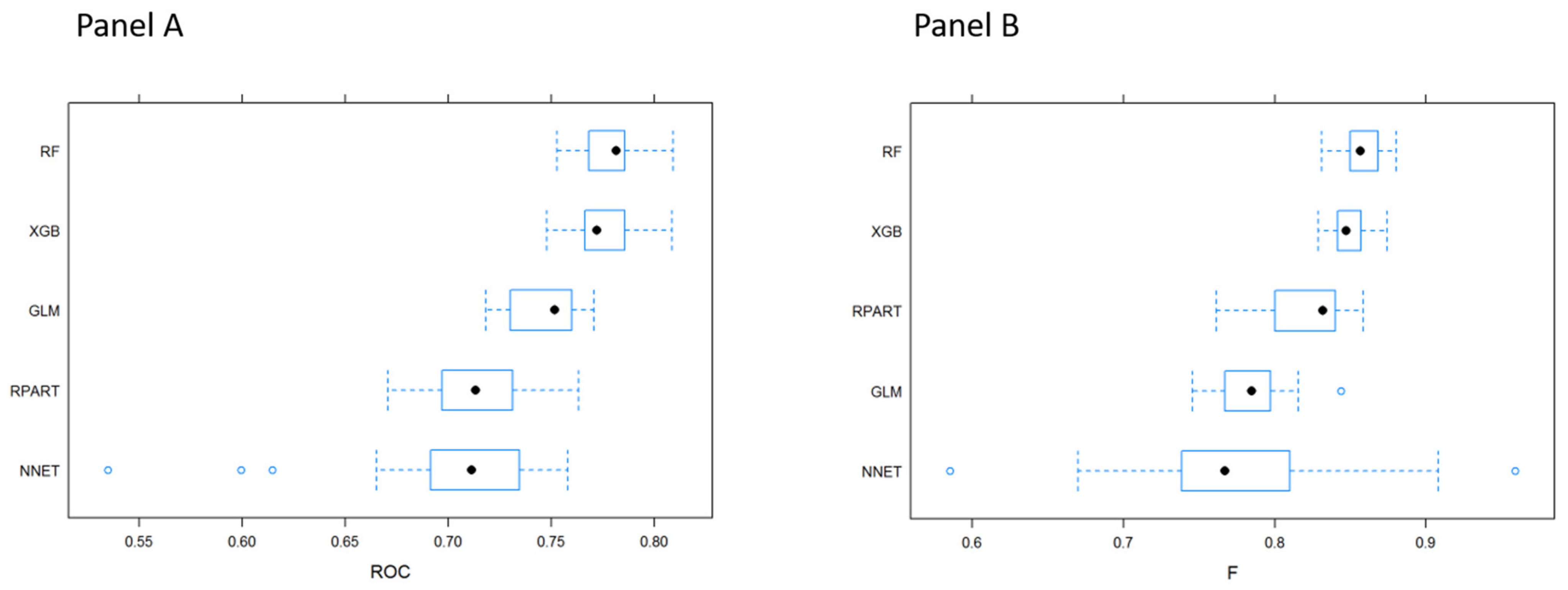
3.1. MLTs Performance
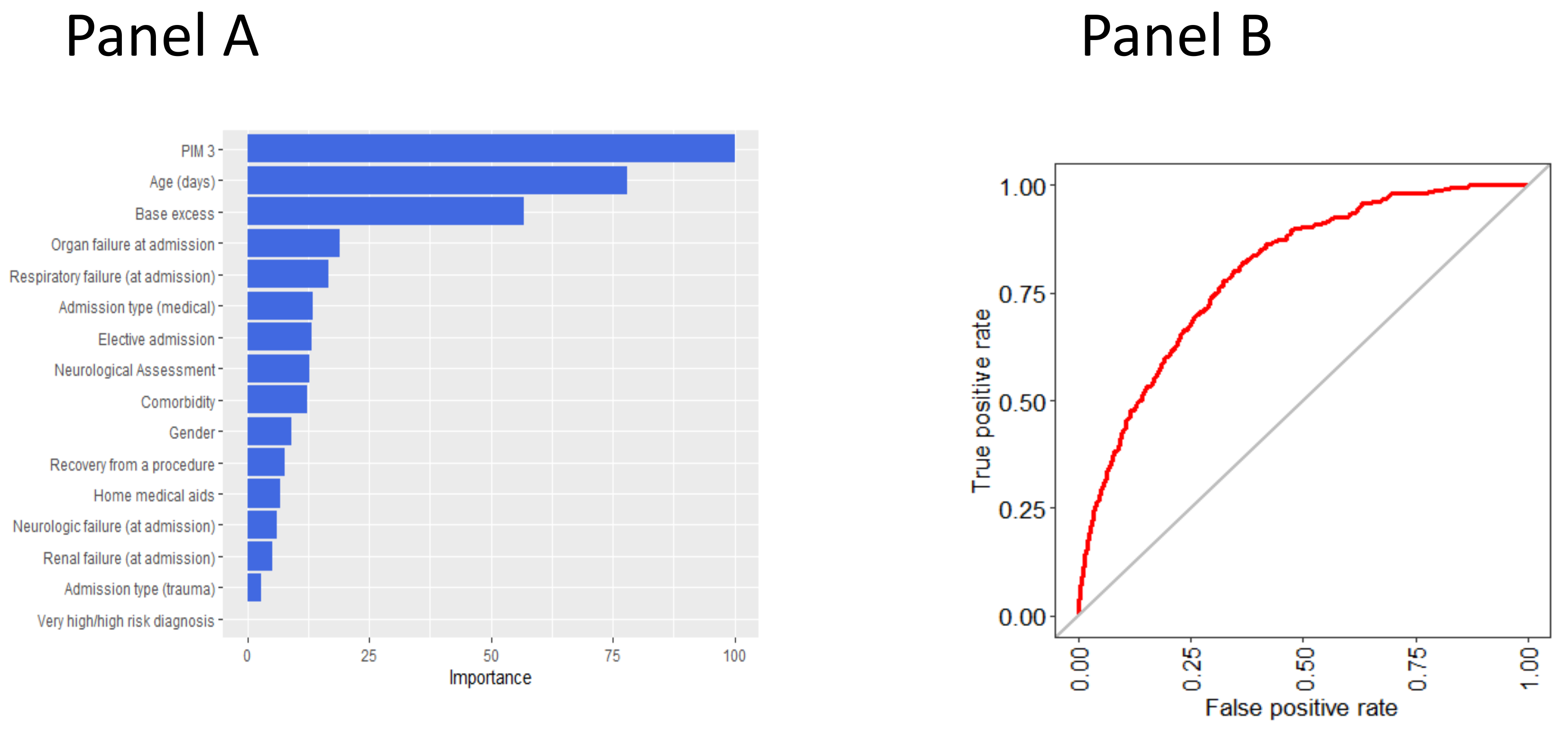
3.2. Variable Importance in Predicting the Outcome According to the Random Forest Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hsu, D.T.; Pearson, G.D. Heart Failure in Children. Circ. Heart Fail. 2009, 2, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaddy, R.E.; George, A.T.; Jaecklin, T.; Lochlainn, E.N.; Thakur, L.; Agrawal, R.; Solar-Yohay, S.; Chen, F.; Rossano, J.W.; Severin, T.; et al. Systematic Literature Review on the Incidence and Prevalence of Heart Failure in Children and Adolescents. Pediatr. Cardiol. 2018, 39, 415–436. [Google Scholar] [CrossRef] [Green Version]
- Multisystem Inflammatory Syndrome in Children and Adolescents Temporally Related to COVID-19. Available online: https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19 (accessed on 21 May 2021).
- Mpanya, D.; Celik, T.; Klug, E.; Ntsinjana, H. Predicting Mortality and Hospitalization in Heart Failure Using Machine Learning: A Systematic Literature Review. Int. J. Cardiol. Heart Vasc. 2021, 34. [Google Scholar] [CrossRef]
- Angraal, S.; Mortazavi, B.J.; Gupta, A.; Khera, R.; Ahmad, T.; Desai, N.R.; Jacoby, D.L.; Masoudi, F.A.; Spertus, J.A.; Krumholz, H.M. Machine Learning Prediction of Mortality and Hospitalization in Heart Failure With Preserved Ejection Fraction. JACC Heart Fail. 2020, 8, 12–21. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. Machine Learning Can Predict Survival of Patients with Heart Failure from Serum Creatinine and Ejection Fraction Alone. BMC Med. Inform. Decis. Mak. 2020, 20. [Google Scholar] [CrossRef]
- Lorenzoni, G.; Sabato, S.S.; Lanera, C.; Bottigliengo, D.; Minto, C.; Ocagli, H.; de Paolis, P.; Gregori, D.; Iliceto, S.; Pisanò, F. Comparison of Machine Learning Techniques for Prediction of Hospitalization in Heart Failure Patients. J. Clin. Med. 2019, 8, 1298. [Google Scholar] [CrossRef] [Green Version]
- Ngiam, K.Y.; Khor, I.W. Big Data and Machine Learning Algorithms for Health-Care Delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef]
- Wu, T.T.; Lin, X.Q.; Mu, Y.; Li, H.; Guo, Y.S. Machine Learning for Early Prediction of In-hospital Cardiac Arrest in Patients with Acute Coronary Syndromes. Clin. Cardiol. 2021, 44, 349–356. [Google Scholar] [CrossRef]
- Spelmen, V.S.; Porkodi, R. A Review on Handling Imbalanced Data. In Proceedings of the 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT), Coimbatore, India, 1–3 March 2018; pp. 1–11. [Google Scholar]
- Solanki, A.; Kumar, S.; Nayyar, A. Handbook of Research on Emerging Trends and Applications of Machine Learning; IGI Global: Hershey, PA, USA, 2020; ISBN 978-1-5231-2888-4. [Google Scholar]
- Straney, L.; Clements, A.; Parslow, R.C.; Pearson, G.; Shann, F.; Alexander, J.; Slater, A. Paediatric Index of Mortality 3: An Updated Model for Predicting Mortality in Pediatric Intensive Care. Pediatr. Crit. Care Med. 2013, 14, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research Electronic Data Capture (REDCap)—A Metadata-Driven Methodology and Workflow Process for Providing Translational Research Informatics Support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models, 2nd ed.; Monographs on Statistics and Applied Probability; Chapman & Hall/CRC: Boca Raton, FL, USA, 1998; ISBN 978-0-412-31760-6. [Google Scholar]
- Breiman, L. (Ed.) Classification and Regression Trees, 1st ed.; Chapman & Hall/CRC: Boca Raton, FL, USA, 1998; ISBN 978-0-412-04841-8. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2009; ISBN 978-0-387-84857-0. [Google Scholar]
- Breiman, L. Random Forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Bishop, C.M. Neural Networks for Pattern Recognition; Clarendon Press: New York, NY, USA, 1995; ISBN 978-0-19-853849-3. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Kuhn, M.; Johnson, K. Remedies for Severe Class Imbalance. In Applied Predictive Modeling; Springer: New York, NY, USA, 2013; pp. 419–443. ISBN 978-1-4614-6848-6. [Google Scholar]
- Ali, A.; Shamsuddin, S.; Ralescu, A. Classification with Class Imbalance Problem: A Review. Int. J. Adv. Soft Comput. Appl. 2013, 5, 1–30. [Google Scholar]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of Imbalanced Data: A Review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Van Buuren, S.; Brand, J.P.L.; Groothuis-Oudshoorn, C.G.M.; Rubin, D.B. Fully Conditional Specification in Multivariate Imputation. J. Stat. Comput. Simul. 2006, 76, 1049–1064. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Efron, B.; Tibshirani, R. An Introduction to the Bootstrap; Monographs on Statistics and Applied Probability; Chapman & Hall: New York, NY, USA, 1993; ISBN 978-0-412-04231-7. [Google Scholar]
- Miao, Y.; Kirby, A.; Boscardin, W.J. Estimating Harrell’s Optimism on Predictive Indices Using Bootstrap Samples. In Proceedings of the SAS Global Forum; University of California San Francisco: San Francisco, CA, USA, 2013; Volume 504. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Harrell, F.E., Jr. RMS: Regression Modeling Strategies; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training [R Package Version 6.0-88]. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 31 May 2021).
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Extreme Gradient Boosting [R Package Xgboost Version 1.2.0.1]. 2020. Available online: https://CRAN.R-project.org/package=xgboost (accessed on 31 May 2021).
- Churpek, M.M.; Yuen, T.C.; Winslow, C.; Meltzer, D.O.; Kattan, M.W.; Edelson, D.P. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit. Care Med. 2016, 44, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Yun, K.; Oh, J.; Hong, T.H.; Kim, E.Y. Prediction of Mortality in Surgical Intensive Care Unit Patients Using Machine Learning Algorithms. Front. Med. 2021, 8. [Google Scholar] [CrossRef]
- Lee, B.; Kim, K.; Hwang, H.; Kim, Y.S.; Chung, E.H.; Yoon, J.-S.; Cho, H.J.; Park, J.D. Development of a Machine Learning Model for Predicting Pediatric Mortality in the Early Stages of Intensive Care Unit Admission. Sci. Rep. 2021, 11, 1263. [Google Scholar] [CrossRef]
- Zhang, D.; Yin, C.; Hunold, K.M.; Jiang, X.; Caterino, J.M.; Zhang, P. An Interpretable Deep-Learning Model for Early Prediction of Sepsis in the Emergency Department. Patterns 2021, 2. [Google Scholar] [CrossRef]
- Zhai, H.; Brady, P.; Li, Q.; Lingren, T.; Ni, Y.; Wheeler, D.S.; Solti, I. Developing and Evaluating a Machine Learning Based Algorithm to Predict the Need of Pediatric Intensive Care Unit Transfer for Newly Hospitalized Children. Resuscitation 2014, 85, 1065–1071. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Kwon, Y.S.; Baek, M.S. Machine Learning Models to Predict 30-Day Mortality in Mechanically Ventilated Patients. J. Clin. Med. 2021, 10, 2172. [Google Scholar] [CrossRef]
- Tezza, F.; Lorenzoni, G.; Azzolina, D.; Barbar, S.; Leone, L.A.C.; Gregori, D. Predicting In-Hospital Mortality of Patients with COVID-19 Using Machine Learning Techniques. J. Pers. Med. 2021, 11, 343. [Google Scholar] [CrossRef]
- Hu, C.-A.; Chen, C.-M.; Fang, Y.-C.; Liang, S.-J.; Wang, H.-C.; Fang, W.-F.; Sheu, C.-C.; Perng, W.-C.; Yang, K.-Y.; Kao, K.-C.; et al. Using a Machine Learning Approach to Predict Mortality in Critically Ill Influenza Patients: A Cross-Sectional Retrospective Multicentre Study in Taiwan. BMJ Open 2020, 10, e033898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.; Davis, D.N. Cluster Based Under-Sampling for Unbalanced Cardiovascular Data. In Proceedings of the Lecture Notes in Engineering and Computer Science, WCE 2013, London, UK, 3–5 July 2013. [Google Scholar]
- Liu, Y.; Yu, X.; Huang, J.X.; An, A. Combining Integrated Sampling with SVM Ensembles for Learning from Imbalanced Datasets. Inf. Process. Manag. 2011, 47, 617–631. [Google Scholar] [CrossRef]
- Rahman, M.M.; Davis, D.N. Addressing the Class Imbalance Problem in Medical Datasets. Int. J. Mach. Learn. Comput. 2013, 224–228. [Google Scholar] [CrossRef]
- Drumnond, C.; Holte, R.C. Class Imbalance and Cost Sensitivity: Why Undersampling Beats Oversampling. In Proceedings of the ICML-KDD 2003 Workshop: Learning from Imbalanced Datasets, Washington, DC, USA, 21 August 2003. [Google Scholar]
| Characteristics | N | No HF (N = 29,095) | HF (N = 399) | Combined (N = 29,494) | p-Value |
|---|---|---|---|---|---|
| Age (days) | 29,494 | 133/754/2766 | 132/881/2874 | 133/755/2768 | 0.402 |
| Gender: Male | 29,494 | 57% (16,559) | 51% (204) | 57% (16,763) | 0.020 |
| Female | 43% (12,536) | 49% (195) | 43% (12,731) | ||
| Newborn: Yes | 29,485 | 6% (1665) | 2% (7) | 6% (1672) | <0.001 |
| No | 94% (27,421) | 98% (392) | 94% (27,813) | ||
| Comorbidity: Yes | 27,931 | 45% (12,451) | 59% (235) | 45% (12,686) | <0.001 |
| No | 55% (15,081) | 41% (164) | 55% (15,245) | ||
| Home medical aids: Yes | 26,153 | 7% (1886) | 5% (20) | 7% (1906) | 0.114 |
| No | 93% (23,883) | 95% (364) | 93% (24,247) | ||
| Admission type: Surgical | 27,921 | 45% (12,322) | 17% (68) | 44% (12,390) | <0.001 |
| Medical | 50% (13,771) | 76% (304) | 50% (14,075) | ||
| Trauma | 5% (1429) | 7% (27) | 5% (1456) | ||
| Organ failure at admission: Yes | 27,458 | 58% (15,759) | 87% (349) | 59% (16,108) | <0.001 |
| No | 42% (11,300) | 13% (50) | 41% (11,350) | ||
| Respiratory failure (at admission): Yes | 29,494 | 48% (14,010) | 76% (305) | 49% (14,315) | <0.001 |
| No | 52% (15,085) | 24% (94) | 51% (15,179) | ||
| Neurologic failure (at admission): Yes | 29,494 | 9% (2538) | 19% (77) | 9% (2615) | <0.001 |
| No | 91% (26,557) | 81% (322) | 91% (26,879) | ||
| Renal failure (at admission): Yes | 29,494 | 3% (823) | 12% (47) | 3% (870) | <0.001 |
| No | 97% (28,272) | 88% (352) | 97% (28,624) | ||
| Drug sedation: Yes | 27,454 | 32% (8686) | 31% (124) | 32% (8810) | 0.663 |
| No | 68% (18,369) | 69% (275) | 68% (18,644) | ||
| Ventilation in the first hour: Yes | 26,393 | 48% (12,378) | 66% (252) | 48% (12,630) | <0.001 |
| No | 52% (13,636) | 34% (127) | 52% (13,763) | ||
| Base excess | 26,368 | −3.0/0.0/0.0 | −4.7/0.0/1.2 | −3.0/0.0/0.0 | 0.143 |
| Elective admission: Yes | 26,390 | 44% (11,351) | 19% (71) | 43% (11,422) | <0.001 |
| No | 56% (14,660) | 81% (308) | 57% (14,968) | ||
| Recovery from a procedure: Yes | 26,394 | 47% (12,159) | 28% (108) | 46% (12,267) | <0.001 |
| No | 53% (13,855) | 72% (272) | 54% (14,127) | ||
| PIM 3 | 26,280 | 0.28/0.77/2.20 | 1.02/3.25/8.12 | 0.28/0.79/2.27 | <0.001 |
| Very high/high risk diagnosis: Yes | 29,494 | 89% (25,764) | 88% (351) | 89% (26,115) | 0.717 |
| No | 11% (3331) | 12% (48) | 11% (3379) |
| Technique | Sampling Method | Sensitivity | Specificity | Accuracy | ROC |
|---|---|---|---|---|---|
| GLM | Original sampling | 1/1/1 | 0/0/0 | 0.985/0.986/0.987 | 0.756/0.769/0.782 |
| Downsampling | 0.672/0.690/0.705 | 0.651/0.681/0.714 | 0.672/0.690/0.703 | 0.745/0.756/0.771 | |
| Upsampling | 0.659/0.671/0.675 | 0.688/0.699/0.735 | 0.659/0.671/0.675 | 0.761/0.769/0.776 | |
| RPART | Original sampling | 0.997/0.998/0.999 | 0/0.0123/0.020 | 0.984/0.985/0.986 | 0.608/0.643/0.709 |
| Downsampling | 0.672/0.715/0.725 | 0.566/0.596/0.669 | 0.673/0.714/0.723 | 0.704/0.719/0.724 | |
| Upsampling | 0.940/0.943/0.946 | 0.171/0.194/0.218 | 0.930/0.933/0.936 | 0.559/0.568/0.580 | |
| NNET | Original sampling | 0.999/0.999/1 | 0/0/0 | 0.985/0.986/0.986 | 0.711/0.722/0.741 |
| Downsampling | 0.585/0.625/0.685 | 0.690/0.762/0.791 | 0.588/0.627/0.685 | 0.694/0.720/0.745 | |
| Upsampling | 0.638/0.669/0.700 | 0.675/0.717/0.757 | 0.639/0.670/0.700 | 0.715/0.725/0.742 | |
| RF | Original sampling | 1/1/1 | 0/0/0 | 0.986/0.986/0.987 | 0.739/0.748/0.757 |
| Downsampling | 0.756/0.763/0.771 | 0.587/0.623/0.640 | 0.754/0.761/0.768 | 0.757/0.769/0.776 | |
| Upsampling | 0.999/1/1 | 0/0.007/0.014 | 0.985/0.985/0.986 | 0.731/0.739/0.744 | |
| XGB | Original sampling | 1/1/1 | 0/0/0.006 | 0.986/0.986/0.987 | 0.750/0.758/0.769 |
| Downsampling | 0.689/0.696/0.714 | 0.673/0.713/0.750 | 0.689/0.695/0.714 | 0.770/0.780/0.793 | |
| Upsampling | 0.962/0.964/0.966 | 0.156/0.171/0.180 | 0.952/0.953/0.954 | 0.718/0.723/0.733 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comoretto, R.I.; Azzolina, D.; Amigoni, A.; Stoppa, G.; Todino, F.; Wolfler, A.; Gregori, D.; on behalf of the TIPNet Study Group. Predicting Hemodynamic Failure Development in PICU Using Machine Learning Techniques. Diagnostics 2021, 11, 1299. https://doi.org/10.3390/diagnostics11071299
Comoretto RI, Azzolina D, Amigoni A, Stoppa G, Todino F, Wolfler A, Gregori D, on behalf of the TIPNet Study Group. Predicting Hemodynamic Failure Development in PICU Using Machine Learning Techniques. Diagnostics. 2021; 11(7):1299. https://doi.org/10.3390/diagnostics11071299
Chicago/Turabian StyleComoretto, Rosanna I., Danila Azzolina, Angela Amigoni, Giorgia Stoppa, Federica Todino, Andrea Wolfler, Dario Gregori, and on behalf of the TIPNet Study Group. 2021. "Predicting Hemodynamic Failure Development in PICU Using Machine Learning Techniques" Diagnostics 11, no. 7: 1299. https://doi.org/10.3390/diagnostics11071299
APA StyleComoretto, R. I., Azzolina, D., Amigoni, A., Stoppa, G., Todino, F., Wolfler, A., Gregori, D., & on behalf of the TIPNet Study Group. (2021). Predicting Hemodynamic Failure Development in PICU Using Machine Learning Techniques. Diagnostics, 11(7), 1299. https://doi.org/10.3390/diagnostics11071299








