Role of Artificial Intelligence in Video Capsule Endoscopy
Abstract
:1. Introduction
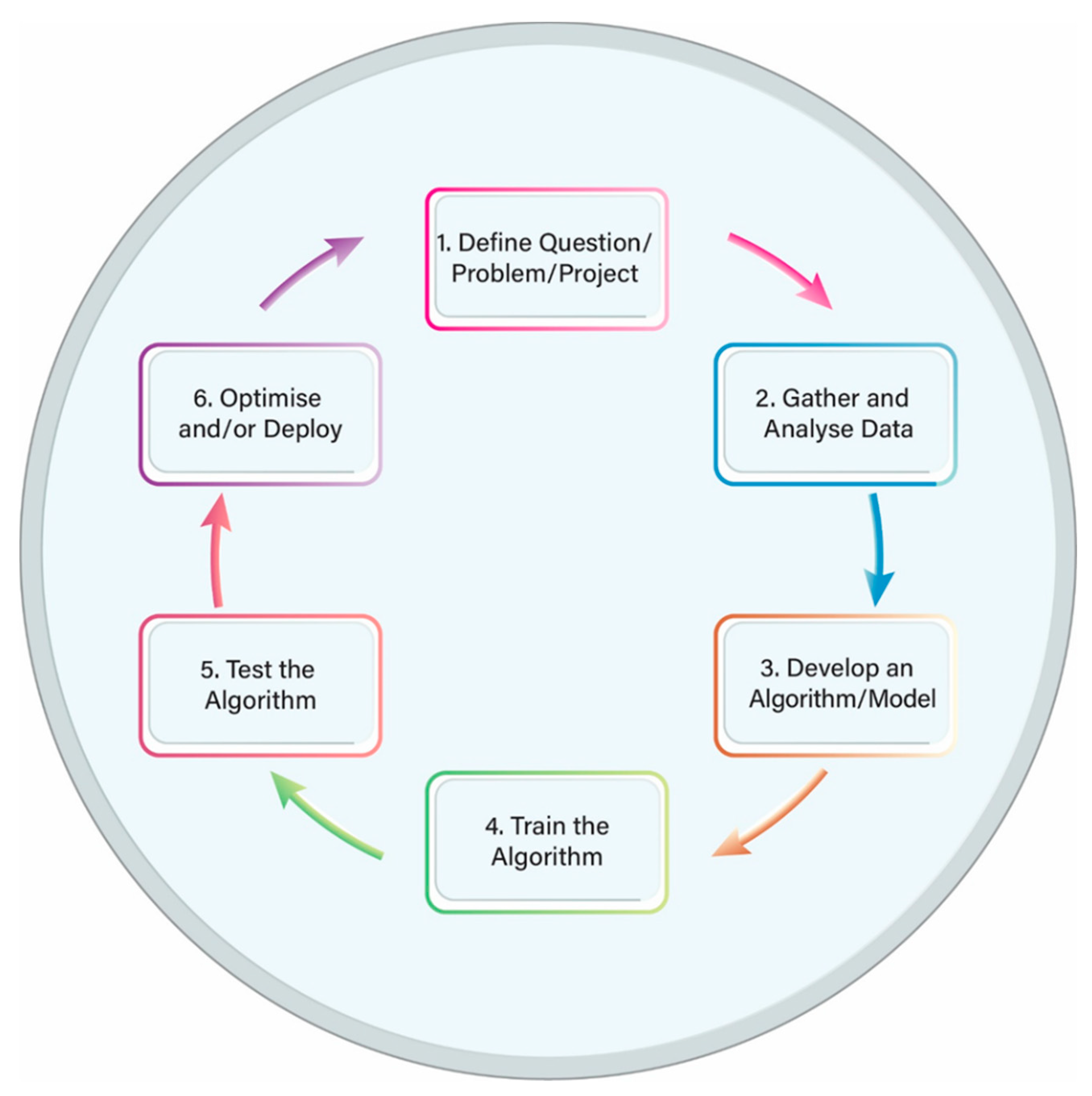
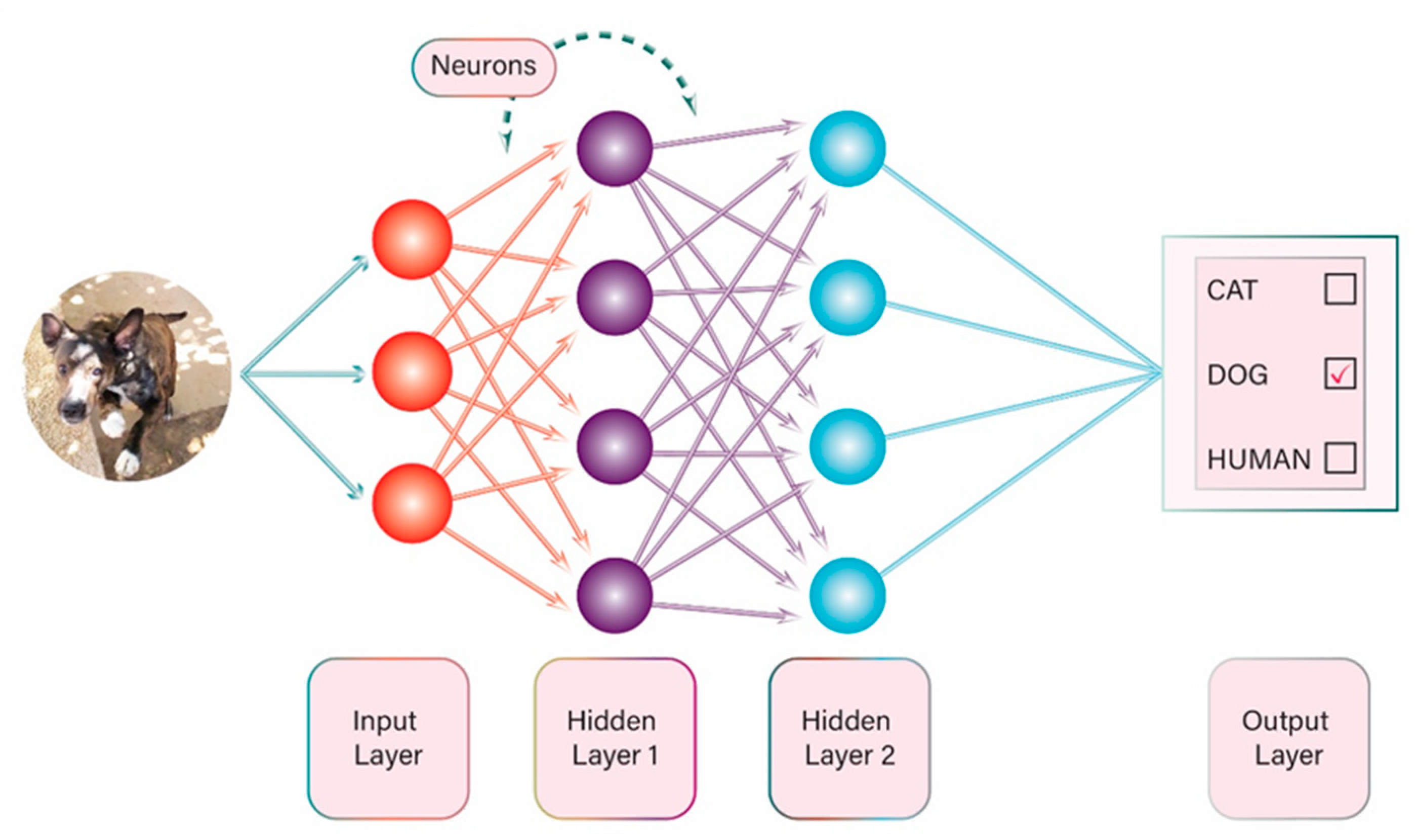
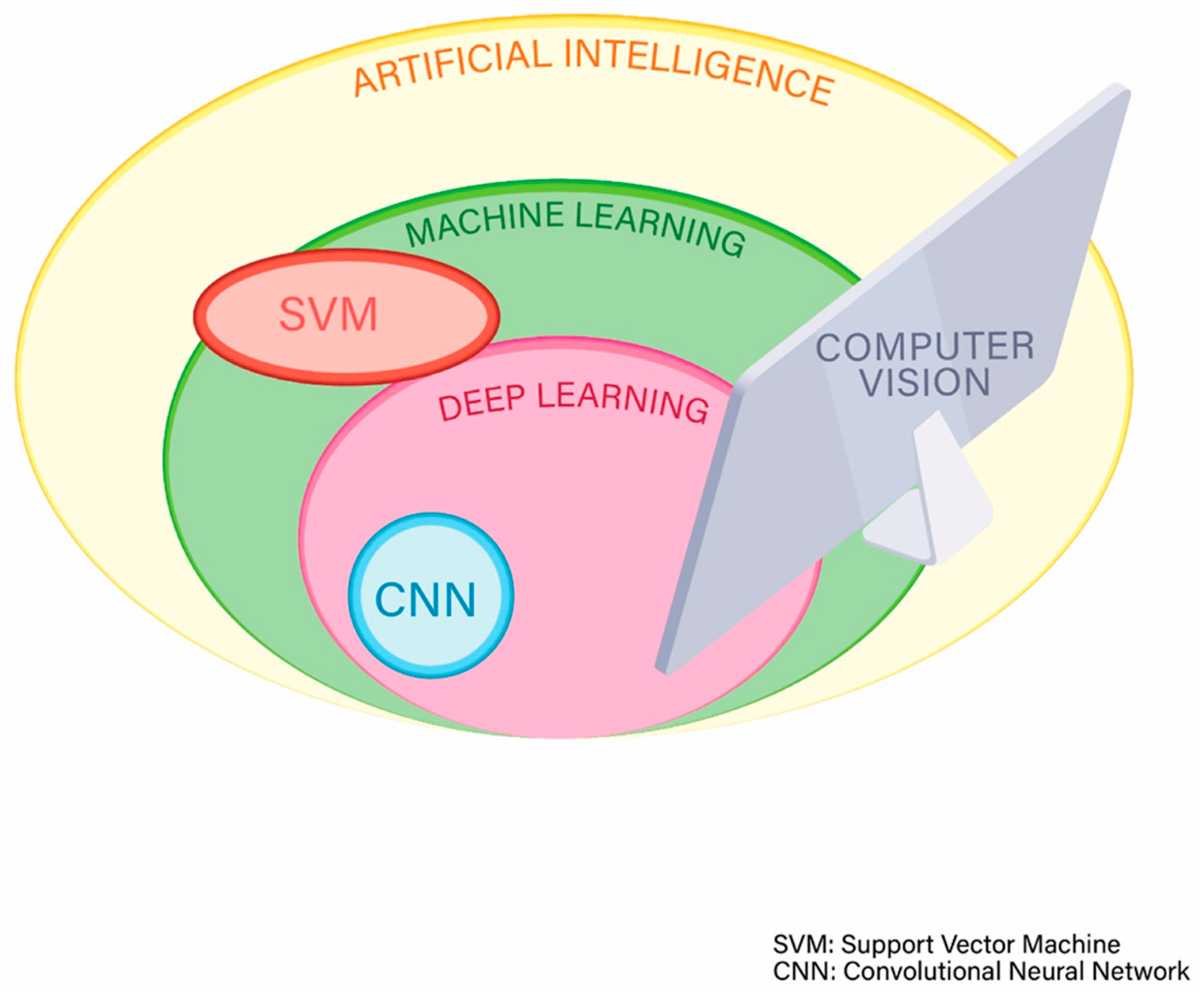
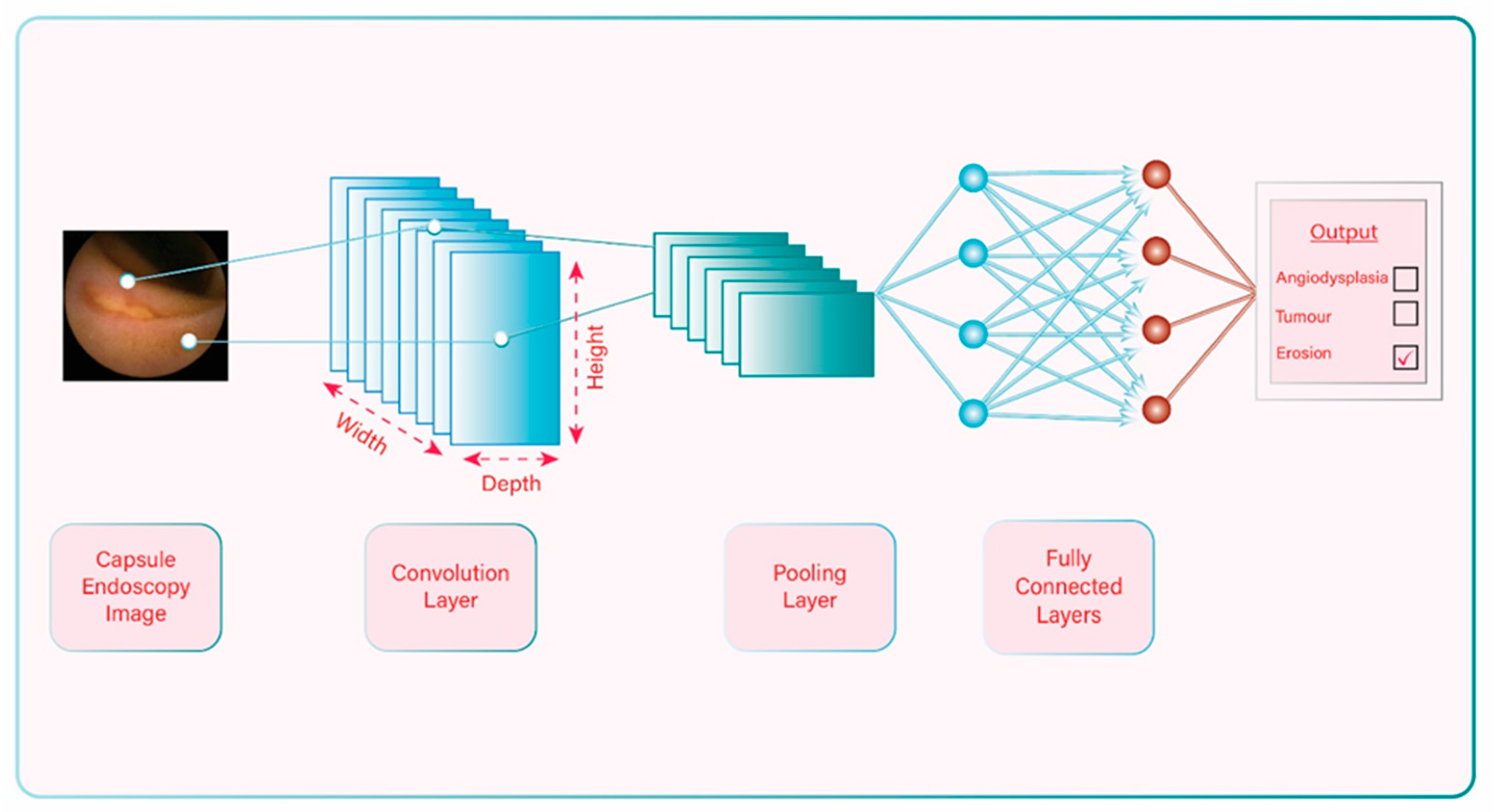
2. Artificial Intelligence: General Information and Terminology
3. Progress of Artificial Intelligence-aided Diagnosis in Capsule Endoscopy and Pathology Detection
3.1. Ulcer/Erosions
3.2. Small Bowel Bleeding and Angioectasias
3.3. Protruding Lesions
3.4. Early Detection of Prolonged Gastric Transit Time
3.5. Celiac Disease
3.6. Intestinal Hookworm Infection
3.7. Multiple Lesion Detection
- Some studies have low numbers of frames/videos (mainly the older studies) and this is limiting the statistical power of the results. Nevertheless, in recent years some centers have created large databases allowing more reliable results in their studies. It seems that we now have enough data available for processing and this is important for future studies.
- Frames with inadequate bowel preparation or reduced image quality were not included in some studies [53] or it is not known if they were included in others. This may affect false-positive or false negative results.
- Only pictures from a single CE system or version were used in most studies [47,59,64,66]. This factor may limit a more widespread application of the algorithm. An algorithm that was designed using images and videos from one CE system may not have the same reliability if used in a database with images from a different CE system or version.
- The large majority of the studies published up to date were designed to assess for specific pathology, e.g., the algorithm detects only blood or ulcers in the small bowel. This is a significant limitation because clinicians are looking for a specific pathology only in a few cases, for example to assess disease activity in a patient with diagnosed Crohn’s disease. More commonly, the etiology of symptoms is unknown, and so it would be extremely useful to have an algorithm that could detect various types of lesions in the small bowel. Some researchers have worked towards this direction.
4. Implementation of Artificial Intelligence in Capsule Endoscopy
- Error reduction due to human limitations such as biases, fatigue or inexperience;
- Improvement of training and learning opportunities: AI technology can be used to provide clinicians with only abnormal CE images for review.
- Cost;
- Need for large databases (CE images or videos). These databases are necessary for training and testing in order to increase sensitivity and specificity and achieve excellent results. It appears that in the last few years, large databases are being increasingly created in many centers around the world and this will not be a problem in the future.
- Is testing of the AI system with images and videos from an existing database adequate or should it be tested on real patient CE images/videos before it comes into practice? Some of the AI algorithms have been checked on real patient data but others have not;
- How can one be sure that a new AI algorithm will have minimal or no problems with overfitting and spectrum bias? These are two of the most common problems that AI engineers have to deal with when designing a new AI algorithm. Overfitting occurs when an algorithm becomes so accurate on a limited dataset that its predictions are not well generalised to new datasets. Most often the algorithm is overfitted to the training dataset. Spectrum bias occurs when the dataset used for the development of the algorithm does not adequately represent the range of disease manifestations or patients that will be encountered in clinical practice (target population) [73];
- What is the gold standard method that a new AI system should be compared to? Is it the conventional reading of CE by expert clinicians? Is it the conventional reading of CE by the clinicians of the hospital where the AI system will be implemented? Possibly, a different AI method will be the gold standard in the future;
- To date, all known studies are retrospective. Large prospective studies are warranted to validate new AI systems in CE. A multicenter, multinational, blinded prospective trial is currently evaluating the role of AI in small bowel capsule endoscopy for obscure GI bleeding (ClinicalTrials.gov Identifier: NCT04821349);
- Cost may be a limitation for widespread use.
- How will the AI system be used in day-to-day practice? For example, a company may have developed an AI system that can detect abnormal CE images with sensitivity of 95% when tested on real patient CE videos. Is the false-negative 5% rate acceptable and should clinicians review only the abnormal images detected by the system (AI-based auxiliary reading)? Conversely, is this sensitivity not acceptable and should clinicians review the whole CE video of patients in order to achieve a higher sensitivity (combination of AI +human reading)?
- How difficult is it to use these new AI systems? How long is the learning curve?
- How can we define malpractice after implementation of CADe/CADx?
5. The Future of Artificial Intelligence-Aided Capsule Endoscopy
- Excellent performance (high sensitivity/specificity/accuracy and very low error rate) in abnormal CE image detection;
- Ability to detect multiple lesions;
- Ability to categorise the lesions;
- Fast reading time of CE videos;
- Easy to use.
- Locomotion [77]. The ability to navigate the capsule within the bowel with the use of an internal or external system is very appealing and could help solve some difficult problems including prolonged capsule transit time in the stomach, insufficient lesion assessment and bowel stenosis. Several companies have expressed interest and have conducted experiments to design and study such a system. At present, the most promising technology uses a magnetic capsule with an external navigation system but is not yet widely implemented.
- Capsule with biopsy properties [81];
- Capsule with improved technological characteristics. The new capsules that will be launched onto the market in the future will offer better quality of images (high resolution) and longer battery life.
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clancey, W.J.; Shortliffe, E.H.; Buchanan, B.G. Intelligent computer-aided instruction for medical diagnosis. Proc. Annu. Symp. Comput. Appl. Med. Care 1979, 175–183. [Google Scholar]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Emmanuel, A.; Patel, M.; Williams, S.; Haji, A.; Hayee, B.; Neumann, H. Artificial intelligence in luminal endoscopy. Ther. Adv. Gastrointest. Endosc. 2020, 13, 2631774520935220. [Google Scholar] [CrossRef]
- Alagappan, M.; Brown, J.R.G.; Mori, Y.; Berzin, T.M. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J. Gastrointest. Endosc. 2018, 10, 239–249. [Google Scholar] [CrossRef]
- Abadir, A.P.; Ali, M.F.; Karnes, W.; Samarasena, J.B. Artificial intelligence in gastrointestinal endoscopy. Clin. Endosc. 2020, 53, 132–141. [Google Scholar] [CrossRef]
- El Hajjar, A.; Rey, J.-F. Artificial intelligence in gastrointestinal endoscopy: General overview. Chin. Med. J. 2020, 133, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, A.; Van Dam, J.; Jakobs, R.; Kudis, V.; Hartmann, D.; Damian, U.; Weickert, U.; Schilling, D.; Riemann, J.F. Computer-assisted colonoscopy (the neoguide endoscopy system): Results of the first human clinical trial (“PACE Study”). Am. J. Gastroenterol. 2007, 102, 261–266. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.-E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy. Ann. Intern. Med. 2018, 169, 357–366. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K.; et al. Artificial intelligence-assisted polyp detection for colonoscopy: Initial experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, A.R.; Haque, M.A. Computer-aided gastrointestinal hemorrhage detection in wireless capsule endoscopy videos. Comput. Methods Programs Biomed. 2015, 122, 341–353. [Google Scholar] [CrossRef]
- Jia, X.; Meng, M.Q.-H. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. In Proceedings of the 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2016; pp. 639–642. [Google Scholar] [CrossRef]
- Kanesaka, T.; Lee, T.-C.; Uedo, N.; Lin, K.-P.; Chen, H.-Z.; Lee, J.-Y.; Wang, H.-P.; Chang, H.-T. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest. Endosc. 2018, 87, 1339–1344. [Google Scholar] [CrossRef]
- Miyaki, R.; Yoshida, S.; Tanaka, S.; Kominami, Y.; Sanomura, Y.; Matsuo, T.; Oka, S.; Raytchev, B.; Tamaki, T.; Koide, T.; et al. A computer system to be used with laser-based endoscopy for quantitative diagnosis of early gastric cancer. J. Clin. Gastroenterol. 2015, 49, 108–115. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, S.; Kim, J.-H.; Keum, J.-S.; Jo, J.; Cha, J.H.; Jung, D.H.; Park, J.J.; Youn, Y.H.; Park, H. Sa1235 application of artificial intelligence for prediction of invasion depth in early gastric cancer: Preliminary study. Gastrointest. Endosc. 2018, 87, AB176. [Google Scholar] [CrossRef]
- Lee, B.-I.; Matsuda, T. Estimation of invasion depth: The first key to successful colorectal ESD. Clin. Endosc. 2019, 52, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Kudo, S.-E.; Chiu, P.; Singh, R.; Misawa, M.; Wakamura, K.; Kudo, T.; Hayashi, T.; Katagiri, A.; Miyachi, H.; et al. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: An international web-based study. Endoscopy 2016, 48, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Nakamura, H.; Kataoka, S.; Maeda, Y.; Kudo, T.; Hayashi, T.; Wakamura, K.; Miyachi, H.; et al. Characterization of Colorectal lesions using a computer-aided diagnostic system for narrow-band imaging endocytoscopy. Gastroenterology 2016, 150, 1531–1532.e3. [Google Scholar] [CrossRef] [Green Version]
- Maeda, Y.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Ogata, N.; Sasanuma, S.; Wakamura, K.; Oda, M.; Mori, K.; Ohtsuka, K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest. Endosc. 2019, 89, 408–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffle, J.K.; Farmer, A.D.; Aziz, Q. Artificial intelligence-assisted gastroenterology—Promises and pitfalls. Am. J. Gastroenterol. 2019, 114, 422–428. [Google Scholar] [CrossRef] [PubMed]
- De Groof, J.; van der Sommen, F.; Van Der Putten, J.; Struyvenberg, M.R.; Zinger, S.; Curvers, W.L.; Pech, O.; Meining, A.; Neuhaus, H.; Bisschops, R.; et al. The Argos project: The development of a computer-aided detection system to improve detection of Barrett’s neoplasia on white light endoscopy. United Eur. Gastroenterol. J. 2019, 7, 538–547. [Google Scholar] [CrossRef] [Green Version]
- Ichimasa, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Matsudaira, S.; Kouyama, Y.; Baba, T.; Hidaka, E.; Wakamura, K.; Hayashi, T.; et al. Artificial intelligence may help in predicting the need for additional surgery after endoscopic resection of T1 colorectal cancer. Endoscopy 2017, 50, 230–240. [Google Scholar] [CrossRef]
- Das, A.; Ben-Menachem, T.; Cooper, G.S.; Chak, A.; Sivak, M.V.; A Gonet, J.; Wong, R.C. Prediction of outcome in acute lower-gastrointestinal haemorrhage based on an artificial neural network: Internal and external validation of a predictive model. Lancet 2003, 362, 1261–1266. [Google Scholar] [CrossRef]
- Adadi, A.; Adadi, S.; Berrada, M. Gastroenterology meets machine learning: Status quo and quo vadis. Adv. Bioinform. 2019, 2019, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Yang, Y.; Wu, F.; Gao, M.; Xu, Y.; Zhang, Y.; Yao, Y.; Du, X.; Li, C.; Wu, L.; et al. Discriminative analysis of schizophrenia using support vector machine and recursive feature elimination on structural MRI images. Medicine 2016, 95, e3973. [Google Scholar] [CrossRef] [PubMed]
- Korfiatis, P.; Kline, T.L.; Coufalova, L.; Lachance, D.H.; Parney, I.F.; Carter, R.E.; Buckner, J.C.; Erickson, B.J. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 2016, 43, 2835–2844. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Cai, H.; Tan, W.; Jin, C.; Li, L. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci. Rep. 2016, 6, 27327. [Google Scholar] [CrossRef] [PubMed]
- Aissa, J.; Schaarschmidt, B.M.; Below, J.; Bethge, O.T.; Böven, J.; Sawicki, L.M.; Hoff, N.-P.; Kröpil, P.; Antoch, G.; Boos, J. Performance and clinical impact of machine learning based lung nodule detection using vessel suppression in melanoma patients. Clin. Imaging 2018, 52, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Vesal, S.; Ravikumar, N.; Ellman, S.; Maier, A. Comparative analysis of unsupervised algorithms for breast MRI lesion segmentation. In Bildverarbeitung für die Medizin; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 257–262. [Google Scholar]
- Xu, J.; Zhou, C.; Lang, B.; Liu, Q. Deep Learning for Histopathological Image Analysis: Towards Computerized Diagnosis on Cancers; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2017; pp. 73–95. [Google Scholar]
- Barry, J.D.; Fagny, M.; Paulson, J.N.; Aerts, H.J.; Platig, J.; Quackenbush, J. Histopathological image QTL Discovery of immune infiltration variants. iScience 2018, 5, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Abdolmanafi, A.; Duong, L.; Dahdah, N.; Cheriet, F. Deep feature learning for automatic tissue classification of coronary artery using optical coherence tomography. Biomed. Opt. Express 2017, 8, 1203–1220. [Google Scholar] [CrossRef] [Green Version]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lee, V.H.F.; Ng, M.K.; Yan, H.; Bijlsma, M.F. Molecular subtyping of cancer: Current status and moving toward clinical applications. Brief. Bioinform. 2018, 20, 572–584. [Google Scholar] [CrossRef]
- Danaee, P.; Ghaeini, R.; Hendrix, D.A. A deep learning approach for cancer detection and relevant gene identification. Biocomputing 2017, 22, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Shao, X.-N.; Sun, Y.-J.; Xiao, K.-T.; Zhang, Y.; Zhang, W.-B.; Kou, Z.-F.; Cheng, J.-L. Texture analysis of magnetic resonance T1 mapping with dilated cardiomyopathy: A machine learning approach. Medicine 2018, 97, e12246. [Google Scholar] [CrossRef]
- Betancur, J.; Otaki, Y.; Motwani, M.; Fish, M.B.; Lemley, M.; Dey, D.; Gransar, H.; Tamarappoo, B.; Germano, G.; Sharir, T.; et al. Prognostic value of combined clinical and myocardial perfusion imaging data using machine learning. JACC Cardiovasc. Imaging 2018, 11, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Rajagopalan, C.; Clifford, G.D. A machine learning approach to multi-level ECG signal quality classification. Comput. Methods Programs Biomed. 2014, 117, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Bouton, C.E.; Shaikhouni, A.; Annetta, N.V.; Bockbrader, M.; Friedenberg, D.A.; Nielson, D.; Sharma, G.; Sederberg, P.B.; Glenn, B.C.; Mysiw, W.J.; et al. Restoring cortical control of functional movement in a human with quadriplegia. Nat. Cell Biol. 2016, 533, 247–250. [Google Scholar] [CrossRef]
- Farina, D.; Vujaklija, I.; Sartori, M.; Kapelner, T.; Negro, F.; Jiang, N.; Bergmeister, K.; Andalib, A.; Principe, J.; Aszmann, O.C. Man/machine interface based on the discharge timings of spinal motor neurons after targeted muscle reinnervation. Nat. Biomed. Eng. 2017, 1, 25. [Google Scholar] [CrossRef] [Green Version]
- Patel, V.; Khan, M.N.; Shrivastava, A.; Sadiq, K.; Ali, S.A.; Moore, S.R.; Brown, D.E.; Syed, S. Artificial intelligence applied to gastrointestinal diagnostics: A review. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019, 13, 388–393. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Shin, K.; Jung, J.; Bae, H.-J.; Kim, D.H.; Byeon, J.-S.; Kim, N. Convolutional neural network technology in endoscopic imaging: Artificial intelligence for endoscopy. Clin. Endosc. 2020, 53, 117–126. [Google Scholar] [CrossRef]
- Park, J.; Hwang, Y.; Yoon, J.-H.; Park, M.-G.; Kim, J.; Lim, Y.J.; Chun, H.J. Recent development of computer vision technology to improve capsule endoscopy. Clin. Endosc. 2019, 52, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2019, 89, 357–363.e2. [Google Scholar] [CrossRef] [PubMed]
- Alaskar, H.; Hussain, A.; Al-Aseem, N.; Liatsis, P.; Al-Jumeily, D. Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images. Sensors 2019, 19, 1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charisis, V.S.; Hadjileontiadis, L.J. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J. Gastroenterol. 2016, 22, 8641–8657. [Google Scholar] [CrossRef]
- Given Imaging. Capsule Endoscopy. 2014. Available online: http://www.capsuleendoscopy.org (accessed on 1 April 2021).
- Iakovidis, D.K.; Koulaouzidis, A. Software for enhanced video capsule endoscopy: Challenges for essential progress. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Koulaouzidis, A.; Iakovidis, D.K.; Yung, D.E.; Rondonotti, E.; Kopylov, U.; Plevris, J.N.; Toth, E.; Eliakim, A.; Johansson, G.W.; Marlicz, W.; et al. KID Project: An internet-based digital video atlas of capsule endoscopy for research purposes. Endosc. Int. Open 2017, 5, E477–E483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klang, E.; Barash, Y.; Margalit, R.Y.; Soffer, S.; Shimon, O.; Albshesh, A.; Ben-Horin, S.; Amitai, M.M.; Eliakim, R.; Kopylov, U. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest. Endosc. 2020, 91, 606–613.e2. [Google Scholar] [CrossRef]
- Kumar, R.; Zhao, Q.; Seshamani, S.; Mullin, G.; Hager, G.; Dassopoulos, T. Assessment of Crohn’s disease lesions in wireless capsule endoscopy images. IEEE Trans. Biomed. Eng. 2011, 59, 355–362. [Google Scholar] [CrossRef]
- Barash, Y.; Azaria, L.; Soffer, S.; Yehuda, R.M.; Shlomi, O.; Ben-Horin, S.; Eliakim, R.; Klang, E.; Kopylov, U. Ulcer severity grading in video capsule images of patients with Crohn’s disease: An ordinal neural network solution. Gastrointest. Endosc. 2021, 93, 187–192. [Google Scholar] [CrossRef]
- Leenhardt, R.; Vasseur, P.; Li, C.; Saurin, J.C.; Rahmi, G.; Cholet, F.; Becq, A.; Marteau, P.; Histace, A.; Dray, X.; et al. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest. Endosc. 2019, 89, 189–194. [Google Scholar] [CrossRef]
- Leenhardt, R.; Li, C.; Le Mouel, J.-P.; Rahmi, G.; Saurin, J.C.; Cholet, F.; Boureille, A.; Amiot, X.; Delvaux, M.; Duburque, C.; et al. CAD-CAP: A 25,000-image database serving the development of artificial intelligence for capsule endoscopy. Endosc. Int. Open 2020, 8, E415–E420. [Google Scholar] [CrossRef] [Green Version]
- Tsuboi, A.; Oka, S.; Aoyama, K.; Saito, H.; Aoki, T.; Yamada, A.; Matsuda, T.; Fujishiro, M.; Ishihara, S.; Nakahori, M.; et al. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig. Endosc. 2020, 32, 382–390. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J. Gastroenterol. Hepatol. 2019, 35, 1196–1200. [Google Scholar] [CrossRef]
- Barbosa, D.C.; Roupar, D.B.; Ramos, J.C.; Tavares, A.C.; Lima, C.S. Automatic small bowel tumor diagnosis by using multi-scale wavelet-based analysis in wireless capsule endoscopy images. Biomed. Eng. Online 2012, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Saito, H.; Aoki, T.; Aoyama, K.; Kato, Y.; Tsuboi, A.; Yamada, A.; Fujishiro, M.; Oka, S.; Ishihara, S.; Matsuda, T.; et al. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2020, 92, 144–151.e1. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.; Liu, S.; Yang, J.; Zeng, B.; Yang, L. A pilot trial of convolution neural network for automatic retention-monitoring of capsule endoscopes in the stomach and duodenal bulb. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Molder, A.; Balaban, D.V.; Jinga, M.; Molder, C.-C. Current Evidence on Computer-Aided Diagnosis of Celiac Disease: Systematic Review. Front. Pharmacol. 2020, 11, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Han, G.; Li, B.N.; Lin, Z.; Ciaccio, E.J.; Green, P.H.; Qin, J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput. Biol. Med. 2017, 85, 1–6. [Google Scholar] [CrossRef] [PubMed]
- He, J.-Y.; Wu, X.; Jiang, Y.-G.; Peng, Q.; Jain, R. Hookworm detection in wireless capsule endoscopy images with deep learning. IEEE Trans. Image Process. 2018, 27, 2379–2392. [Google Scholar] [CrossRef]
- Iakovidis, D.K.; Koulaouzidis, A. Automatic lesion detection in capsule endoscopy based on color saliency: Closer to an essential adjunct for reviewing software. Gastrointest. Endosc. 2014, 80, 877–883. [Google Scholar] [CrossRef]
- Nawarathna, R.; Oh, J.; Muthukudage, J.; Tavanapong, W.; Wong, J.; de Groen, P.C.; Tang, S.J. Abnormal image detection in endoscopy videos using a filter bank and local binary patterns. Neurocomputing 2014, 144, 70–91. [Google Scholar] [CrossRef] [Green Version]
- Iakovidis, D.K.; Georgakopoulos, S.V.; Vasilakakis, M.; Koulaouzidis, A.; Plagianakos, V.P. Detecting and locating gastrointestinal anomalies using deep learning and iterative cluster unification. IEEE Trans. Med. Imaging 2018, 37, 2196–2210. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Shi, H.; Zhang, H.; Meng, L.; Fan, M.; Han, C.; Zhang, K.; Ming, F.; Xie, X.; Liu, H.; et al. Gastroenterologist-Level Identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology 2019, 157, 1044–1054.e5. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Fujisawa, G.; Odawara, N.; Kondo, R.; Tsuboi, A.; Ishibashi, R.; Nakada, A.; et al. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig. Endosc. 2020, 32, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Kominami, Y.; Yoshida, S.; Tanaka, S.; Sanomura, Y.; Hirakawa, T.; Raytchev, B.; Tamaki, T.; Koide, T.; Kaneda, K.; Chayama, K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest. Endosc. 2016, 83, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep learning localizes and identifies polyps in Real Time with 96% accuracy in screening colonoscopy. Gastroenterology 2018, 155, 1069–1078.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Bang, C.S. Application of artificial intelligence in gastroenterology. World J. Gastroenterol. 2019, 25, 1666–1683. [Google Scholar] [CrossRef]
- Slawinski, P.R.; Obstein, K.L.; Valdastri, P. Capsule endoscopy of the future: What’s on the horizon? World J. Gastroenterol. 2015, 21, 10528–10541. [Google Scholar] [CrossRef]
- Vasilakakis, M.D.; Koulaouzidis, A.; Marlicz, W.; Iakovidis, D.K. The future of capsule endoscopy in clinical practice: From diagnostic to therapeutic experimental prototype capsules. Gastroenterol. Rev. 2020, 15, 179–193. [Google Scholar] [CrossRef]
- Yang, Y.J. The future of capsule endoscopy: The role of artificial intelligence and other technical advancements. Clin. Endosc. 2020, 53, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.J.; Kim, K.S.; Lim, Y.J. A New Active Locomotion capsule endoscopy under magnetic control and automated reading program. Clin. Endosc. 2020, 53, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Oumrani, S.; Histace, A.; Ali, E.A.; Pietri, O.; Becq, A.; Houist, G.; Nion-Larmurier, I.; Camus, M.; Florent, C.; Dray, X. Multi-criterion, automated, high-performance, rapid tool for assessing mucosal visualization quality of still images in small bowel capsule endoscopy. Endosc. Int. Open 2019, 7, E944–E948. [Google Scholar] [CrossRef] [Green Version]
- Ali, E.A.; Histace, A.; Camus, M.; Gerometta, R.; Becq, A.; Pietri, O.; Nion-Larmurier, I.; Li, C.; Chaput, U.; Marteau, P.; et al. Development and validation of a computed assessment of cleansing score for evaluation of quality of small-bowel visualization in capsule endoscopy. Endosc. Int. Open 2018, 6, E646–E651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becq, A.; Histace, A.; Camus, M.; Nion-Larmurier, I.; Ali, E.A.; Pietri, O.; Romain, O.; Chaput, U.; Li, C.; Marteau, P.; et al. Development of a computed cleansing score to assess quality of bowel preparation in colon capsule endoscopy. Endosc. Int. Open 2018, 6, E844–E850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, D.; Dogan, M.D.; Sitti, M. Magnetically actuated soft capsule endoscope for fine-needle aspiration biopsy. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Singapore, 29 May–3 June 2017; IEEE: Piscataway, NJ, USA; pp. 1132–1139. [Google Scholar]
- Stewart, F.R.; Newton, I.P.; Nathke, I.; Huang, Z.; Cox, B.F. Development of a therapeutic capsule endoscope for treatment in the gastrointestinal tract: Bench testing to translational trial. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS), Washington, DC, USA, 6–9 September 2017. [Google Scholar] [CrossRef] [Green Version]
- Leung, B.H.K.; Poon, C.C.Y.; Zhang, R.; Zheng, Y.; Chan, C.K.W.; Chiu, P.; Lau, J.Y.W.; Sung, J.J.Y. A therapeutic wireless capsule for treatment of gastrointestinal haemorrhage by balloon tamponade effect. IEEE Trans. Biomed. Eng. 2016, 64, 1106–1114. [Google Scholar] [CrossRef]
| AI System Categories | Areas of Assistance |
|---|---|
| Technical |
|
| Detection (CADe) | |
| Diagnostic (CADx) | |
| Therapeutic |
| Medicine Domain | ML Applications | References |
|---|---|---|
| Radiology | Radiological imaging tasks such as:
| [25,26,27,28,29] |
| Pathology | Digital pathological image analysis notably:
| [30,31,32] |
| Oncology | Early cancer diagnosis and prognosis:
| [33,34,35] |
| Cardiology | Early detection of cardiovascular diseases based on:
| [36,37,38] |
| Neurology | Neurological disorders identification and prediction:
| [39,40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tziortziotis, I.; Laskaratos, F.-M.; Coda, S. Role of Artificial Intelligence in Video Capsule Endoscopy. Diagnostics 2021, 11, 1192. https://doi.org/10.3390/diagnostics11071192
Tziortziotis I, Laskaratos F-M, Coda S. Role of Artificial Intelligence in Video Capsule Endoscopy. Diagnostics. 2021; 11(7):1192. https://doi.org/10.3390/diagnostics11071192
Chicago/Turabian StyleTziortziotis, Ioannis, Faidon-Marios Laskaratos, and Sergio Coda. 2021. "Role of Artificial Intelligence in Video Capsule Endoscopy" Diagnostics 11, no. 7: 1192. https://doi.org/10.3390/diagnostics11071192
APA StyleTziortziotis, I., Laskaratos, F.-M., & Coda, S. (2021). Role of Artificial Intelligence in Video Capsule Endoscopy. Diagnostics, 11(7), 1192. https://doi.org/10.3390/diagnostics11071192







