Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel
Abstract
:1. Introduction
2. Materials and Methods
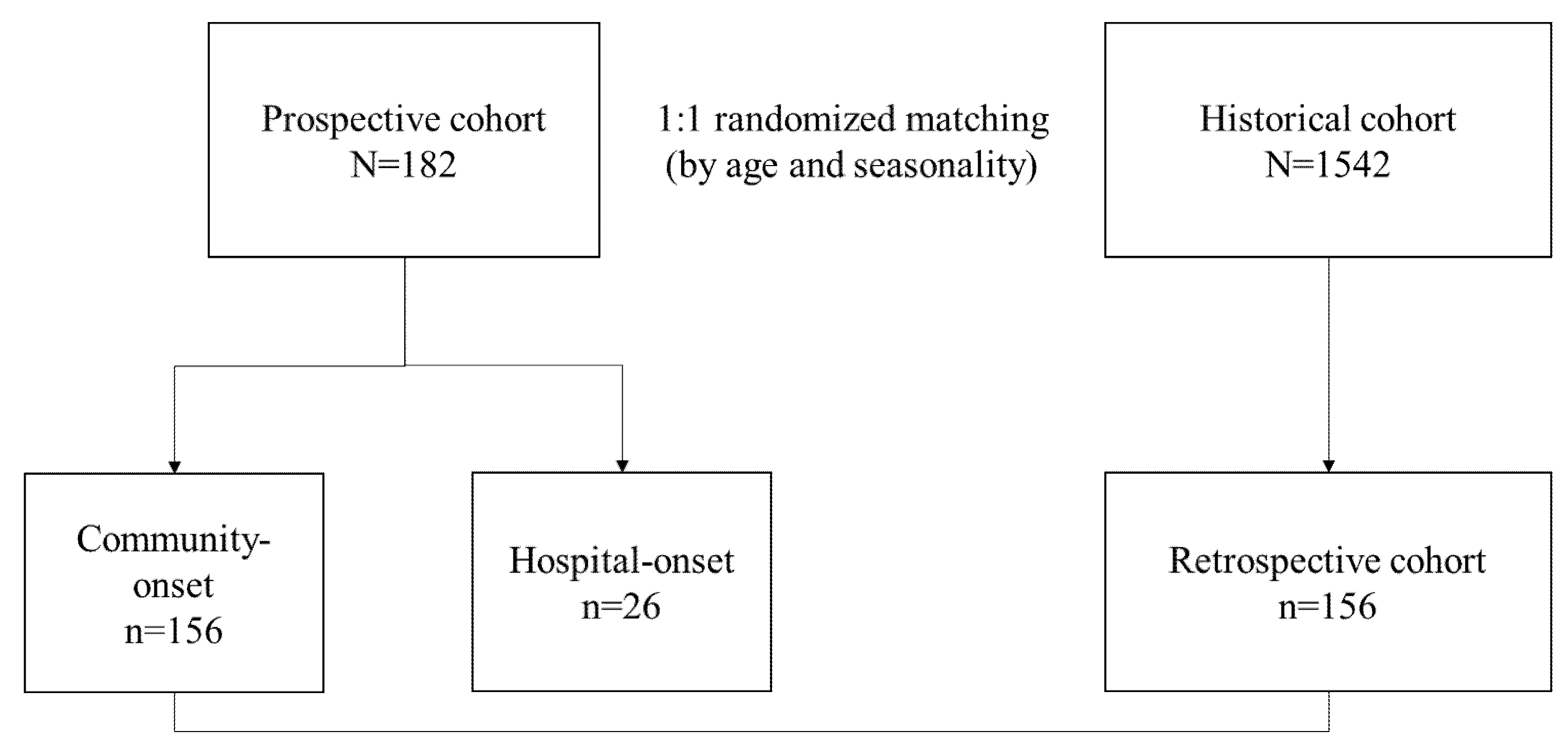
2.1. Study Design and Participants
2.2. Prospective Stool Collection Design
2.2.1. BioFire® FilmArray® Gastrointestinal Panel
2.2.2. Routine Stool Pathogen Detection in the Prospective Cohort
2.3. Retrospective Historical Cohort Study
Stool Pathogen Detection in the Historical Cohort
2.4. Clinical Data Collection
2.5. Statistics
3. Results
3.1. Study Population and Demographics
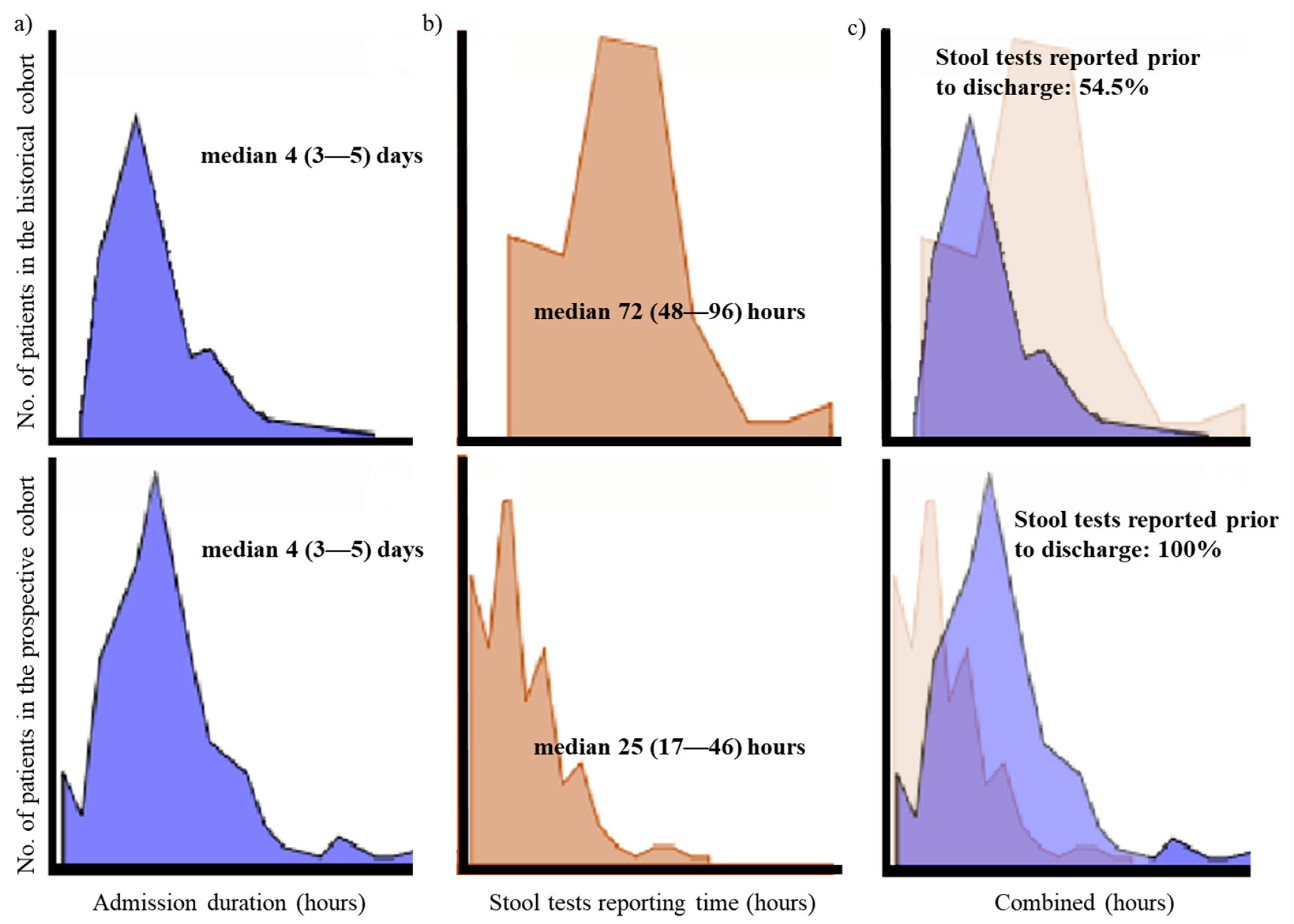
3.2. Comparison of Clinical Parameters between the Prospective and Historical Cohort with Community-Onset Diarrhea
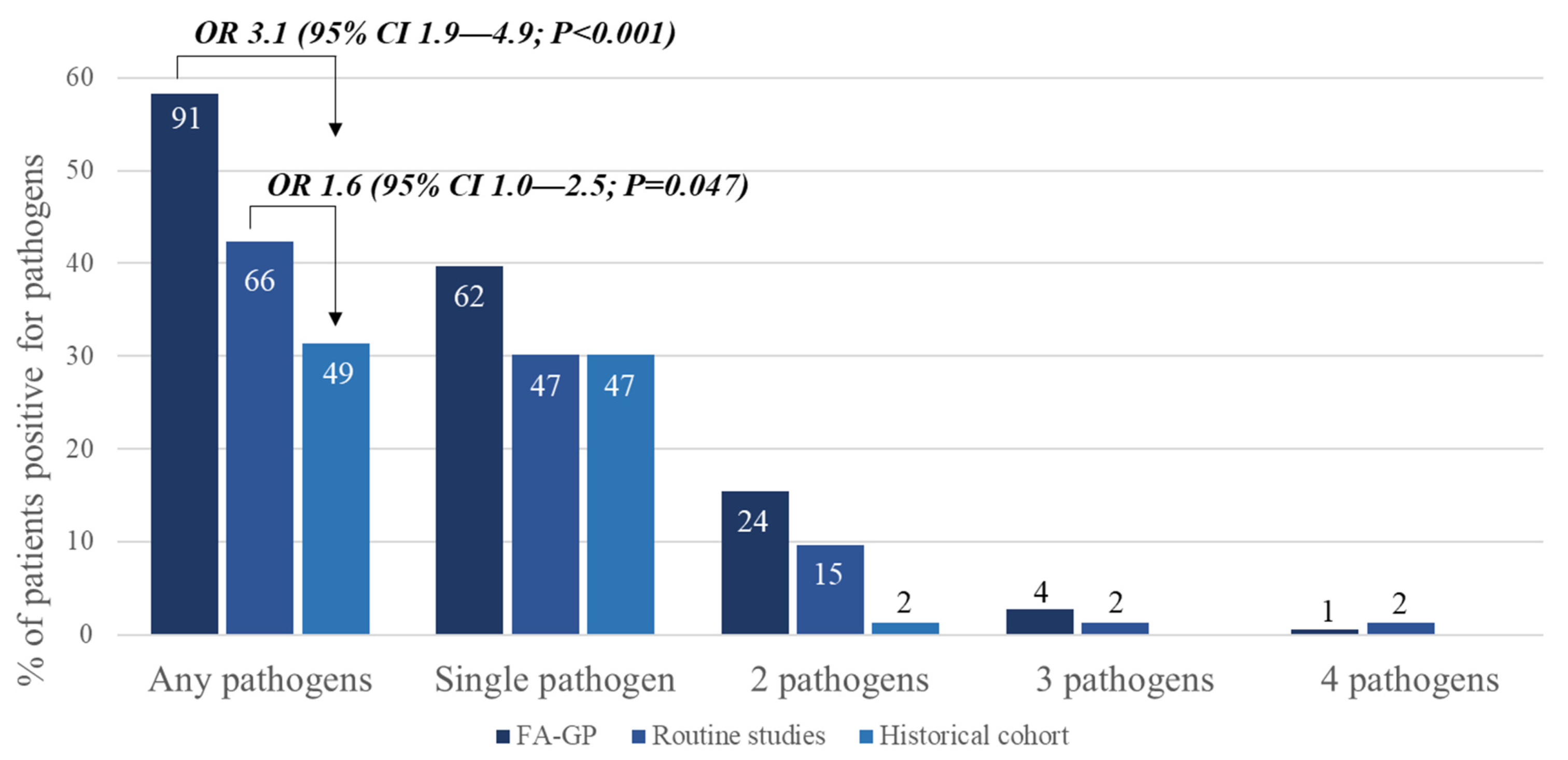
3.3. Comparing BioFire® GI Panel and Routine Conventional Studies in the Prospective Group with Community-Onset Diarrhea
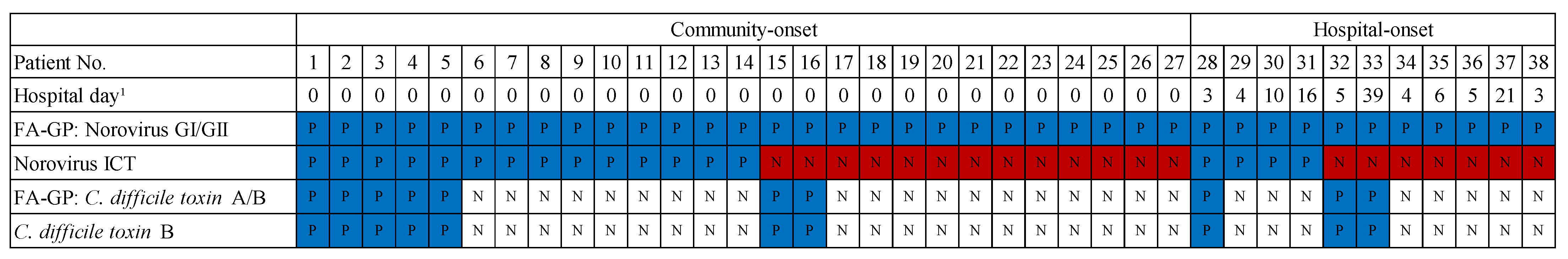
3.4. Utility of the BioFire® GI Panel on Infection Control and Prevention Actions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farthing, M.; Salam, M.A.; Lindberg, G.; Dite, P.; Khalif, I.; Salazar-Lindo, E.; Ramakrishna, B.S.; Goh, K.L.; Thomson, A.; Khan, A.G.; et al. Acute diarrhea in adults and children: A global perspective. J. Clin. Gastroenterol. 2013, 47, 12–20. [Google Scholar] [CrossRef] [Green Version]
- GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef] [Green Version]
- Kosek, M.; Bern, C.; Guerrant, R.L. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ. 2003, 81, 197–204. [Google Scholar] [PubMed]
- Myer, P.A.; Mannalithara, A.; Singh, G.; Singh, G.; Pasricha, P.J.; Ladabaum, U. Clinical and economic burden of emergency department visits due to gastrointestinal diseases in the United States. Am. J. Gastroenterol. 2013, 108, 1496–1507. [Google Scholar] [CrossRef]
- Lo Vecchio, A.; Dias, J.A.; Berkley, J.A.; Boey, C.; Cohen, M.B.; Cruchet, S.; Liguoro, I.; Salazar Lindo, E.; Sandhu, B.; Sherman, P.; et al. Comparison of Recommendations in Clinical Practice Guidelines for Acute Gastroenteritis in Children. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 226–235. [Google Scholar] [CrossRef]
- Cohen, M.B. Etiology and mechanisms of acute infectious diarrhea in infants in the United States. J. Pediatr. 1991, 118, S34–S39. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, K.H.; Park, D.A.; Park, J.; Bang, B.W.; Lee, S.S.; Lee, E.J.; Lee, H.J.; Hong, S.K.; Kim, Y.R. Guideline for the Antibiotic Use in Acute Gastroenteritis. Infect. Chemother. 2019, 51, 217–243. [Google Scholar] [CrossRef] [PubMed]
- Riddle, M.S.; Connor, B.A.; Beeching, N.J.; DuPont, H.L.; Hamer, D.H.; Kozarsky, P.; Libman, M.; Steffen, R.; Taylor, D.; Tribble, D.R.; et al. Guidelines for the prevention and treatment of travelers’ diarrhea: A graded expert panel report. J. Travel Med. 2017, 24, S57–S74. [Google Scholar] [CrossRef] [PubMed]
- Guerrant, R.L.; Van Gilder, T.; Steiner, T.S.; Thielman, N.M.; Slutsker, L.; Tauxe, R.V.; Hennessy, T.; Griffin, P.M.; DuPont, H.; Sack, R.B.; et al. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 2001, 32, 331–351. [Google Scholar] [CrossRef] [PubMed]
- Bruyand, M.; Mariani-Kurkdjian, P.; Gouali, M.; de Valk, H.; King, L.A.; Le Hello, S.; Bonacorsi, S.; Loirat, C. Hemolytic uremic syndrome due to Shiga toxin-producing Escherichia coli infection. Med. Mal. Infect. 2018, 48, 167–174. [Google Scholar] [CrossRef]
- Fraenkel, C.J.; Inghammar, M.; Soderlund-Strand, A.; Johansson, P.J.H.; Bottiger, B. Risk factors for hospital norovirus outbreaks: Impact of vomiting, genotype, and multi-occupancy rooms. J. Hosp. Infect. 2018, 98, 398–403. [Google Scholar] [CrossRef]
- Bennett, W.E., Jr.; Tarr, P.I. Enteric infections and diagnostic testing. Curr. Opin. Gastroenterol. 2009, 25, 1–7. [Google Scholar] [CrossRef]
- Hennessy, T.W.; Marcus, R.; Deneen, V.; Reddy, S.; Vugia, D.; Townes, J.; Bardsley, M.; Swerdlow, D.; Angulo, F.J.; Emerging Infections Program FoodNet Working Group. Survey of physician diagnostic practices for patients with acute diarrhea: Clinical and public health implications. Clin. Infect. Dis. 2004, 38, S203–S211. [Google Scholar] [CrossRef]
- Guarino, A.; Giannattasio, A. New molecular approaches in the diagnosis of acute diarrhea: Advantages for clinicians and researchers. Curr. Opin. Gastroenterol. 2011, 27, 24–29. [Google Scholar] [CrossRef]
- Halligan, E.; Edgeworth, J.; Bisnauthsing, K.; Bible, J.; Cliff, P.; Aarons, E.; Klein, J.; Patel, A.; Goldenberg, S. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: A comparative diagnostic and clinical utility study. Clin. Microbiol. Infect. 2014, 20, O460–O467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- US Food and Drug Administration. Substantial Equivalence Determination Decision Summary. Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/k140407.pdf (accessed on 6 May 2021).
- Torres-Miranda, D.; Akselrod, H.; Karsner, R.; Secco, A.; Silva-Cantillo, D.; Siegel, M.O.; Roberts, A.D.; Simon, G.L. Use of BioFire FilmArray gastrointestinal PCR panel associated with reductions in antibiotic use, time to optimal antibiotics, and length of stay. BMC Gastroenterol. 2020, 20, 246. [Google Scholar] [CrossRef] [PubMed]
- Piralla, A.; Lunghi, G.; Ardissino, G.; Girello, A.; Premoli, M.; Bava, E.; Arghittu, M.; Colombo, M.R.; Cognetto, A.; Bono, P.; et al. FilmArray GI panel performance for the diagnosis of acute gastroenteritis or hemorragic diarrhea. BMC Microbiol. 2017, 17, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buss, S.N.; Leber, A.; Chapin, K.; Fey, P.D.; Bankowski, M.J.; Jones, M.K.; Rogatcheva, M.; Kanack, K.J.; Bourzac, K.M. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J. Clin. Microbiol. 2015, 53, 915–925. [Google Scholar] [CrossRef] [Green Version]
- Machiels, J.D.; Cremers, A.J.H.; van Bergen-Verkuyten, M.; Paardekoper-Strijbosch, S.J.M.; Frijns, K.C.J.; Wertheim, H.F.L.; Rahamat-Langendoen, J.; Melchers, W.J.G. Impact of the BioFire FilmArray gastrointestinal panel on patient care and infection control. PLoS ONE 2020, 15, e0228596. [Google Scholar] [CrossRef]
- Antikainen, J.; Tarkka, E.; Haukka, K.; Siitonen, A.; Vaara, M.; Kirveskari, J. New 16-plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2009, 28, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Khare, R.; Espy, M.J.; Cebelinski, E.; Boxrud, D.; Sloan, L.M.; Cunningham, S.A.; Pritt, B.S.; Patel, R.; Binnicker, M.J. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J. Clin. Microbiol. 2014, 52, 3667–3673. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.S.; Johnson, C.L.; Pritchard, L.; Hepler, R.; Ton, T.T.; Dunn, J.J. Performance of the Verigene(R) enteric pathogens test, Biofire FilmArray gastrointestinal panel and Luminex xTAG(R) gastrointestinal pathogen panel for detection of common enteric pathogens. Diagn. Microbiol. Infect. Dis. 2016, 86, 336–339. [Google Scholar] [CrossRef]
- Axelrad, J.E.; Freedberg, D.E.; Whittier, S.; Greendyke, W.; Lebwohl, B.; Green, D.A. Impact of Gastrointestinal Panel Implementation on Health Care Utilization and Outcomes. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cybulski, R.J., Jr.; Bateman, A.C.; Bourassa, L.; Bryan, A.; Beail, B.; Matsumoto, J.; Cookson, B.T.; Fang, F.C. Clinical Impact of a Multiplex Gastrointestinal Polymerase Chain Reaction Panel in Patients With Acute Gastroenteritis. Clin. Infect. Dis. 2018, 67, 1688–1696. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, Y.S.; Ha, D.J.; Chun, M.J.; Kwon, Y.S. Epidemiology of Rotavirus Gastroenteritis and Rotavirus-Associated Benign Convulsions with Mild Gastroenteritis after the Introduction of Rotavirus Vaccines in South Korea: Nationwide Data from the Health Insurance Review and Assessment Service. Int. J. Environ. Res. Public Health 2020, 17, 8374. [Google Scholar] [CrossRef] [PubMed]
- McKee, R.S.; Schnadower, D.; Tarr, P.I.; Xie, J.; Finkelstein, Y.; Desai, N.; Lane, R.D.; Bergmann, K.R.; Kaplan, R.L.; Hariharan, S.; et al. Predicting Hemolytic Uremic Syndrome and Renal Replacement Therapy in Shiga Toxin-producing Escherichia coli-infected Children. Clin. Infect. Dis. 2020, 70, 1643–1651. [Google Scholar] [CrossRef]
- Han, M.S.; Chung, S.M.; Kim, E.J.; Lee, C.J.; Yun, K.W.; Choe, P.G.; Kim, N.J.; Choi, E.H. Successful control of norovirus outbreak in a pediatric ward with multi-bed rooms. Am. J. Infect. Control 2020, 48, 297–303. [Google Scholar] [CrossRef]
- Jangi, S.; Lamont, J.T. Asymptomatic colonization by Clostridium difficile in infants: Implications for disease in later life. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Enoch, D.A.; Butler, M.J.; Pai, S.; Aliyu, S.H.; Karas, J.A. Clostridium difficile in children: Colonisation and disease. J. Infect. 2011, 63, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Del Vecchio, M.; Alvarez-Uria, A.; Marin, M.; Alcala, L.; Martin, A.; Montilla, P.; Bouza, E. Clinical Significance of Clostridium difficile in Children Less Than 2 Years Old: A Case-Control Study. Pediatr. Infect. Dis. J. 2016, 35, 281–285. [Google Scholar] [CrossRef]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, 987–994. [Google Scholar] [CrossRef] [PubMed]
| N = 182 | |
|---|---|
| Median age, years (IQR) | 3.8 (0.9–8.2) |
| Sex, male | 117 (64.3%) |
| Underlying disease | |
| None | 177 (97.3%) |
| Congenital heart disease | 1 (0.5%) |
| Feeding disorder | 1 (0.5%) |
| Neurologic disease | 2 (1.1%) |
| Hemato-oncologic malignancy | 1 (0.5%) |
| Onset of diarrhea | |
| Community-onset | 156 (85.7%) |
| Hospital-onset | 26 (14.3%) |
| Diagnosed season | |
| Spring | 12 (6.6%) |
| Summer | 57 (31.3%) |
| Fall | 11 (6.0%) |
| Winter | 102 (56.0%) |
| Pathogen detection | |
| Any pathogen | 104 (57.1%) |
| Single pathogen | 71 (39.0%) |
| 2 pathogens | 27 (14.8%) |
| 3 pathogens | 6 (3.3%) |
| Prospective n = 156 | Historical n = 156 | p | |
|---|---|---|---|
| Age, years, median (IQR) | 4.8 (1.2–9.0) | 4.2 (1.0–8.5) | 0.328 |
| Male, n (%) | 98 (62.8) | 95 (61.0) | 0.727 |
| No. of patients admitted, n (%) | 149 (95.5) | 151 (96.8) | 0.556 |
| Season of diagnosis | 1.000 | ||
| Spring, n (%) | 10 (6.4) | 10 (6.4) | |
| Summer, n (%) | 55 (35.3) | 55 (35.3) | |
| Fall, n (%) | 10 (6.4) | 10 (6.4) | |
| Winter, n (%) | 81 (51.9) | 81 (51.9) | |
| WBC, 106/L, median (IQR) | 9110 (6310–12,800) | 9860 (7403–12,918) | 0.124 |
| CRP, mg/dL, median (IQR) | 2.3 (0.2–5.5) | 1.68 (0.32–5.71) | 0.596 |
| Admission duration, days, median (IQR) | 4 (3–5) | 4 (3–5) | 0.079 |
| Stool tests reporting time, hours, median (IQR) | 25 (17–46) | 72 (48–96) | <0.001 |
| No. of Pathogens (%) | p | |||
|---|---|---|---|---|
| BioFire® GI Panel (n = 126) | Historical (n = 51) | |||
| Bacteria | Aeromonas spp. | |||
| Campylobacter spp. | 21 (16.7) | 10 (18.9) | ||
| Clostridium difficile toxin | 19 (15.1) | 4 (7.5) | ||
| Plesiomonas shigelloides | 1 (0.8) | - | ||
| Salmonella spp. | 8 (6.3) | 12 (22.6) | ||
| Yersinia enterocolitica | 6 (4.8) | - | ||
| Yersinia frederiksenii | ||||
| Yersinia pseudotuberculosis | ||||
| Subtotal | 55 (43.7) | 26 (49.1) | 0.375 | |
| Diarrheagenic E. coli/Shigella | EAEC | 1 (0.8) | 4 (7.5) | |
| EIEC | 1 (0.8) | - | ||
| EPEC | 20 (15.9) | 3 (5.7) | ||
| ETEC | 1 (0.8) | 1 (1.9) | ||
| STEC | 4 (3.2) | - | ||
| Subtotal | 27 (21.4) | 8 (15.1) | 0.385 | |
| Virus | Adenovirus | 2 (1.6) | - | |
| Astrovirus | 2 (1.6) | - | ||
| Norovirus | 27 (21.4) | 5 (9.4) | ||
| Rotavirus | 10 (0.8) | 12 (22.6) | ||
| Sapovirus | 3 (2.4) | - | ||
| Subtotal | 44 (34.9) | 17 (32.1) | 0.841 | |
| BioFire® GI Panel | Routine Test | No. of Cases (%) N = 56 |
|---|---|---|
| Negative | Aeromonas spp. | 3 (5.4) |
| Negative | Salmonella spp. | 1 (1.8) |
| Negative | EAEC | 2 (3.6) |
| Negative | Y. frederiksenii | 1 (1.8) |
| Negative | Y. pseudotuberculosis | 1 (1.8) |
| Negative | Rotavirus | 2 (3.6) |
| Subtotal | 10 (17.9) | |
| Clostridium difficile toxin | Negative | 3 (5.4) |
| Plesiomonas shigelloides | Negative | 1 (1.8) |
| Salmonella spp. | Negative | 2 (3.6) |
| EAEC | Negative | 1 (1.8) |
| EIEC | Negative | 1 (1.8) |
| EPEC | Negative | 8 (14.3) |
| STEC | Negative | 2 (3.6) |
| Adenovirus | Negative | 2 (3.6) |
| Astrovirus | Negative | 2 (3.6) |
| Norovirus | Negative | 13 (23.2) |
| Rotavirus | Negative | 8 (14.3) |
| Sapovirus | Negative | 3 (5.4) |
| Subtotal | 46 (82.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, I.H.; Kang, H.M.; Suh, W.; Cho, H.; Yoo, I.Y.; Jo, S.J.; Park, Y.J.; Jeong, D.C. Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel. Diagnostics 2021, 11, 1175. https://doi.org/10.3390/diagnostics11071175
Yoo IH, Kang HM, Suh W, Cho H, Yoo IY, Jo SJ, Park YJ, Jeong DC. Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel. Diagnostics. 2021; 11(7):1175. https://doi.org/10.3390/diagnostics11071175
Chicago/Turabian StyleYoo, In Hyuk, Hyun Mi Kang, Woosuk Suh, Hanwool Cho, In Young Yoo, Sung Jin Jo, Yeon Joon Park, and Dae Chul Jeong. 2021. "Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel" Diagnostics 11, no. 7: 1175. https://doi.org/10.3390/diagnostics11071175
APA StyleYoo, I. H., Kang, H. M., Suh, W., Cho, H., Yoo, I. Y., Jo, S. J., Park, Y. J., & Jeong, D. C. (2021). Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel. Diagnostics, 11(7), 1175. https://doi.org/10.3390/diagnostics11071175







