FSHD1 Diagnosis in a Russian Population Using a qPCR-Based Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples
2.2. The D4Z4 Array Analysis by Blotting
2.3. The D4Z4 Array Analysis by Molecular Combing
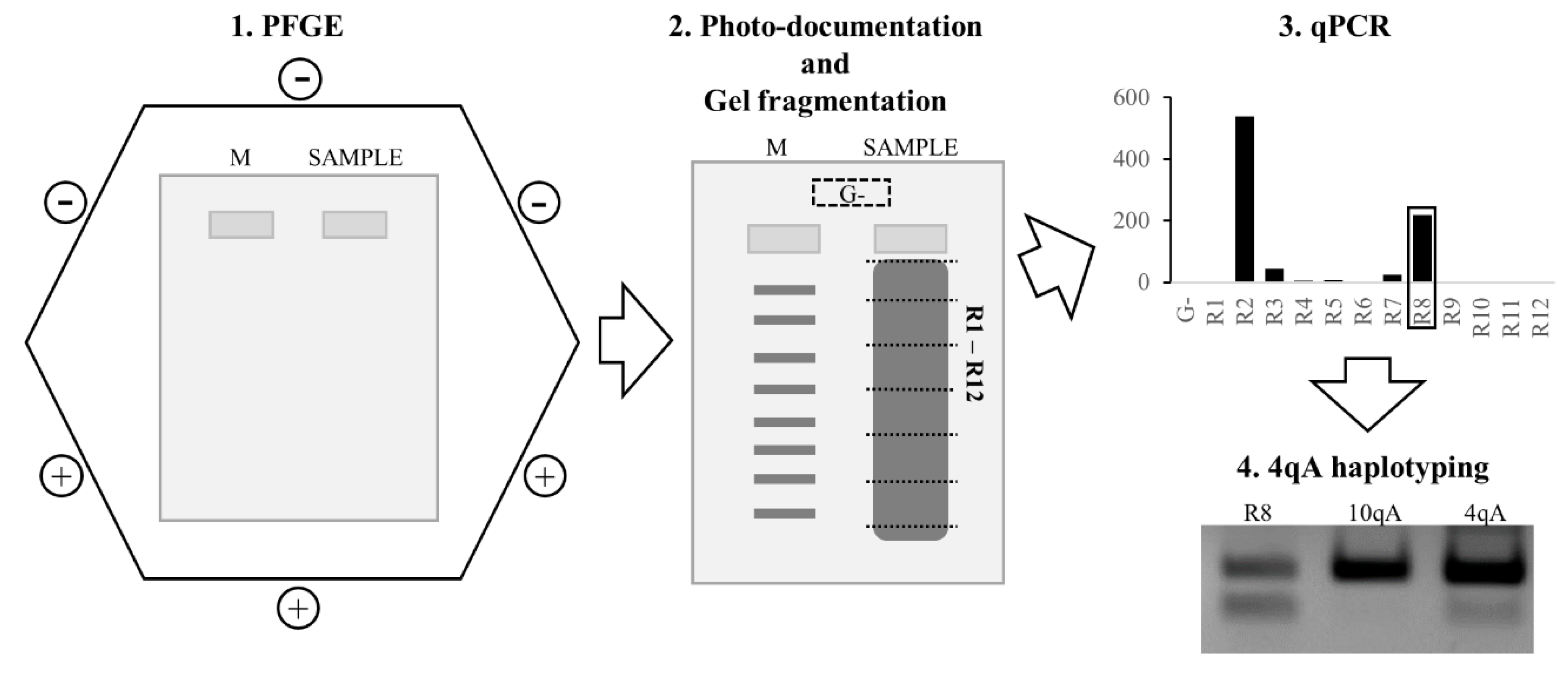
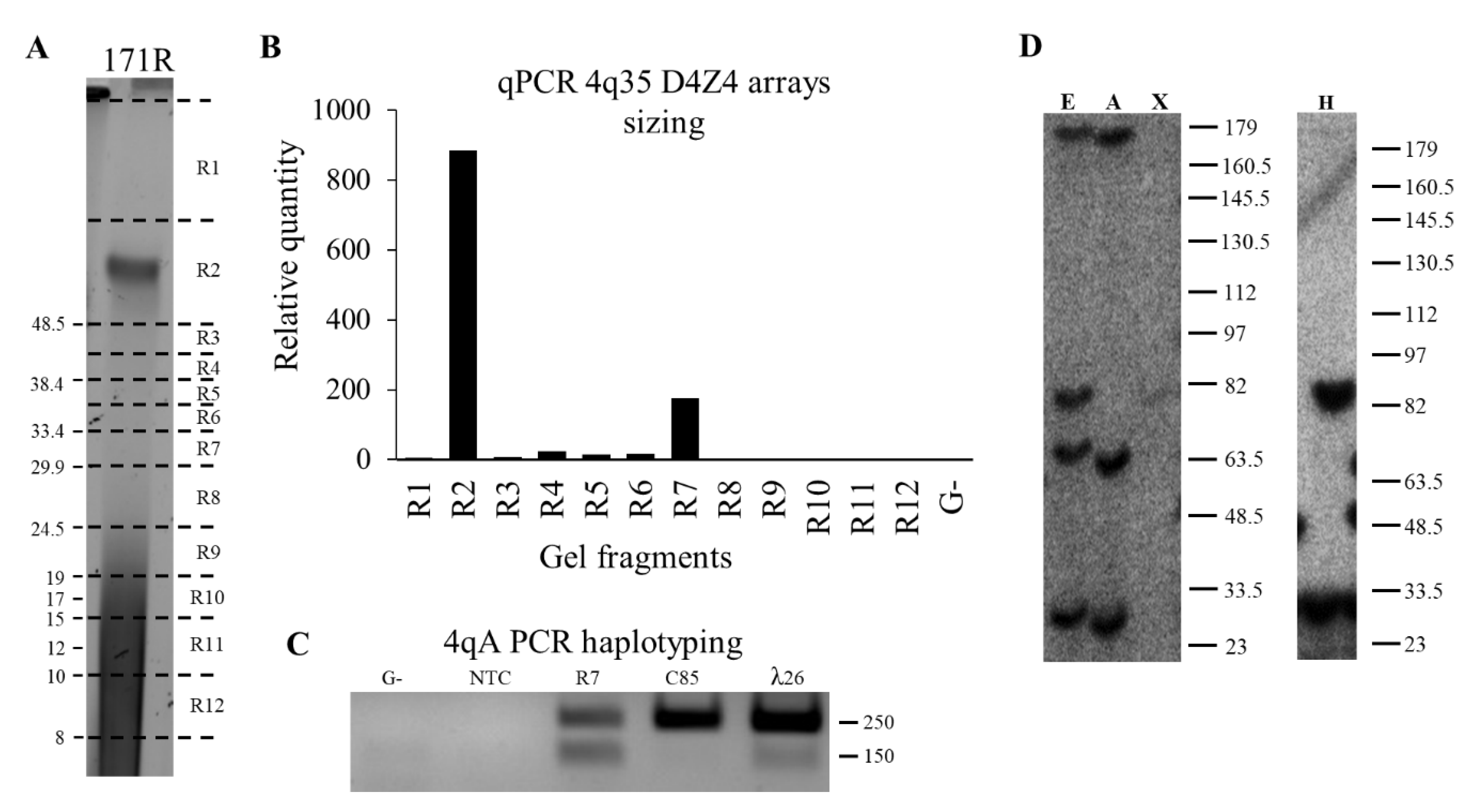
2.4. qPCR-Based D4Z4 Arrays Length Estimation
2.4.1. Pulsed-Field Gel Electrophoresis
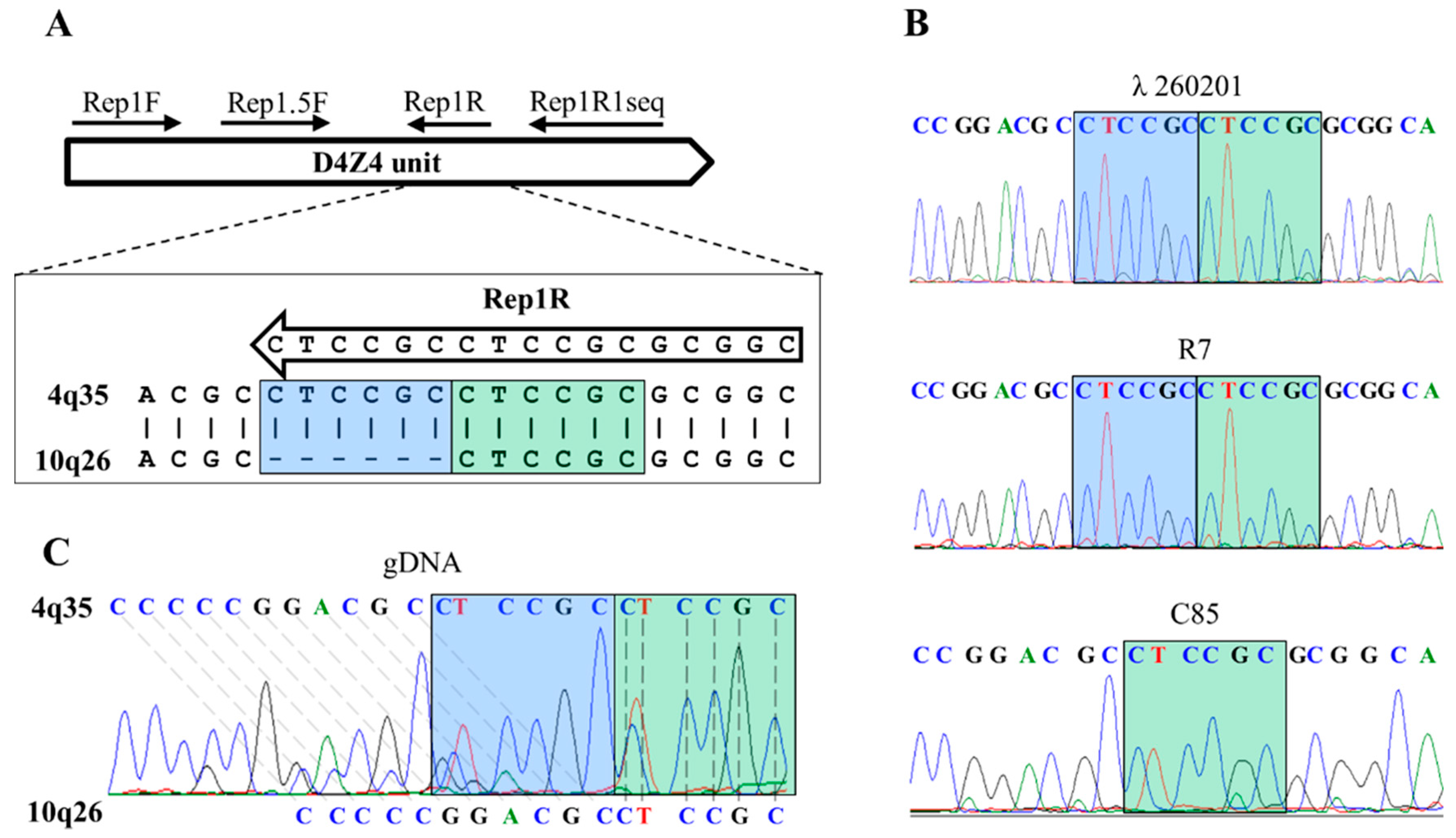
2.4.2. Primers Design
2.4.3. PCR for Sequencing
2.4.4. qPCR-System
2.5. PCR-Haplotyping
2.6. Statistical Analysis
3. Results
3.1. qPCR System for 4q35 D4Z4 Array Length Estimation
3.2. Comparison of the qPCR-Based 4q35 D4Z4 Array Length Estimation against Blotting and Molecular Combing Results
3.3. Comparison of 4qA PCR-Haplotyping Blotting and Molecular Combing Results
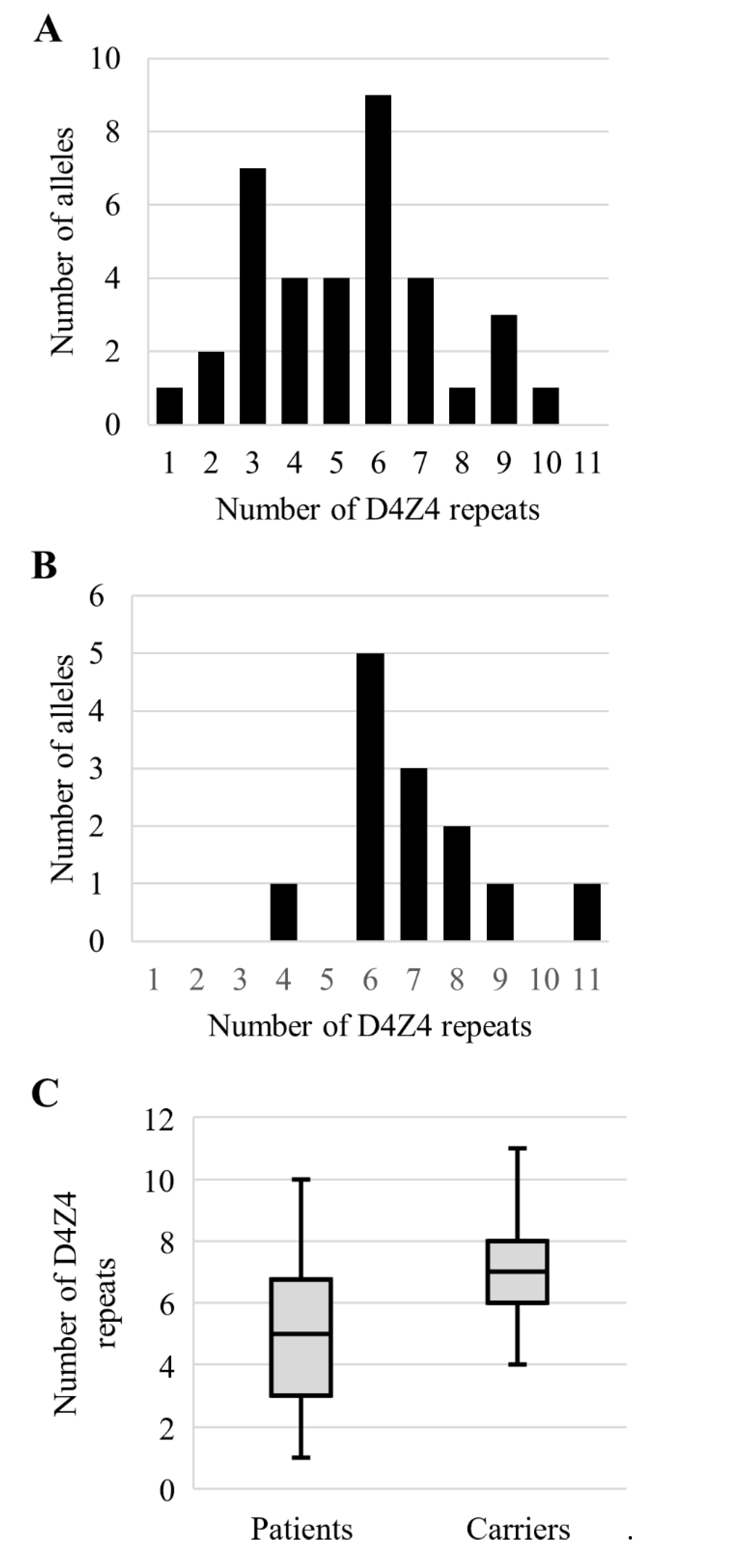
3.4. Analysis of the Permissive Alleles Distribution in the Russian Population
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wijmenga, C.; Van Deutekom, J.C.; Hewitt, J.E.; Padberg, G.W.; Van Ommen, G.-J.B.; Hofker, M.H.; Frants, R.R. Pulsed-Field Gel Electrophoresis of the D4F104S1 Locus Reveals the Size and the Parental Origin of the Facioscapulohumeral Muscular Dystrophy (FSHD)-Associated Deletions. Genomics 1994, 19, 21–26. [Google Scholar] [CrossRef]
- Lunt, P. 44th ENMC International Workshop: Facioscapulohumeral Muscular Dystrophy: Molecular Studies. Neuromuscul. Disord. 1998, 8, 126–130. [Google Scholar] [CrossRef]
- Deidda, G.; Cacurri, S.; Grisanti, P.; Vigneti, E.; Piazzo, N.; Felicetti, L. Physical Mapping Evidence for a Duplicated Region on Chromosome 10qter Showing High Homology with the Facioscapulohumeral Muscular Dystrophy Locus on Chromosome 4qter. Eur. J. Hum. Genet. 1995, 3, 155–167. [Google Scholar] [CrossRef] [PubMed]
- van Deutekom, J.C.; Wijmenga, C.; van Tienhoven, E.A.; Gruter, A.M.; Hewitt, J.E.; Padberg, G.W.; van Ommen, G.J.; Hofker, M.H.; Frants, R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993, 2, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Snider, L.; Geng, L.N.; Lemmers, R.J.L.F.; Kyba, M.; Ware, C.B.; Nelson, A.M.; Tawil, R.; Filippova, G.N.; van der Maarel, S.; Tapscott, S.J.; et al. Facioscapulohumeral Dystrophy: Incomplete Suppression of a Retrotransposed Gene. PLoS Genet. 2010, 6, e1001181. [Google Scholar] [CrossRef] [PubMed]
- Kowaljow, V.; Marcowycz, A.; Ansseau, E.; Conde, C.B.; Sauvage, S.; Mattéotti, C.; Arias, C.; Corona, E.D.; Nuñez, N.G.; Leo, O.; et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007, 17, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Snider, L.; Asawachaicharn, A.; Tyler, A.E.; Geng, L.N.; Petek, L.M.; Maves, L.; Miller, D.; Lemmers, R.J.; Winokur, S.T.; Tawil, R.; et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: New candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009, 18, 2414–2430. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.N.; Yao, Z.; Snider, L.; Fong, A.P.; Cech, J.N.; Young, J.M.; van der Maarel, S.; Ruzzo, W.L.; Gentleman, R.C.; Tawil, R.; et al. DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell 2012, 22, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Dixit, M.; Ansseau, E.; Tassin, A.; Winokur, S.; Shi, R.; Qian, H.; Sauvage, S.; Matteotti, C.; van Acker, A.M.; Leo, O.; et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA 2007, 104, 18157–18162. [Google Scholar] [CrossRef]
- Lemmers, R.J.; De Kievit, P.; Sandkuijl, L.; Padberg, G.W.; Van Ommen, G.-J.B.; Frants, R.R.; van der Maarel, S. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002, 32, 235–236. [Google Scholar] [CrossRef]
- van Geel, M.; Dickson, M.C.; Beck, A.F.; Bolland, D.J.; Frants, R.R.; van der Maarel, S.; de Jong, P.J.; Hewitt, J.E. Genomic Analysis of Human Chromosome 10q and 4q Telomeres Suggests a Common Origin. Genomics 2002, 79, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Mostacciuolo, M.L.; Pastorello, E.; Vazza, G.; Miorin, M.; Angelini, C.; Tomelleri, G.; Galluzzi, G.; Trevisan, C.P. Facioscapulohumeral muscular dystrophy: Epidemiological and molecular study in a north-east Italian population sample. Clin. Genet. 2009, 75, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Wijmenga, C.; Hewitt, J.E.; Sandkuijl, L.A.; Clark, L.N.; Wright, T.J.; Dauwerse, H.G.; Gruter, A.-M.; Hofker, M.H.; Moerer, P.; Williamson, R.; et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992, 2, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Statland, J.M.; Tawil, R. Facioscapulohumeral Muscular Dystrophy. Contin. Lifelong Learn. Neurol. 2016, 22, 1916–1931. [Google Scholar] [CrossRef] [PubMed]
- Lemmers, R.J.L.F.; Tawil, R.; Petek, L.M.; Balog, J.; Block, G.J.; Santen, G.W.E.; Santen, G.W.; Amell, A.M.; Van Der Vliet, P.J.; Almomani, R.; et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012, 44, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Lemmers, R.J.L.F.; Van Der Vliet, P.J.; Klooster, R.; Sacconi, S.; Camaño, P.; Dauwerse, J.G.; Snider, L.; Straasheijm, K.R.; Van Ommen, G.J.; Padberg, G.W.; et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science 2010, 329, 1650–1653. [Google Scholar] [CrossRef]
- Lemmers, R. Analyzing Copy Number Variation Using Pulsed-Field Gel Electrophoresis: Providing a Genetic Diagnosis for FSHD1. In Genotyping: Methods and Protocols; White, S.J., Cantsilieris, S., Eds.; Springer Science+Business Media: New York, NY, USA, 2017; Volume 1492, pp. 107–125. [Google Scholar]
- Nguyen, K.; Walrafen, P.; Bernard, R.; Attarian, S.; Chaix, C.; Vovan, C.; Renard, E.; Dufrane, N.; Pouget, J.; Vannier, A.; et al. Molecular combing reveals allelic combinations in facioscapulohumeral dystrophy. Ann. Neurol. 2011, 70, 627–633. [Google Scholar] [CrossRef]
- Nguyen, K.; Puppo, F.; Roche, S.; Gaillard, M.-C.; Chaix, C.; Lagarde, A.; Pierret, M.; Vovan, C.; Olschwang, S.; Salort-Campana, E.; et al. Molecular combing reveals complex 4q35 rearrangements in Facioscapulohumeral dystrophy. Hum. Mutat. 2017, 38, 1432–1441. [Google Scholar] [CrossRef]
- Wright, T.J.; Wijmenga, C.; Clark, L.N.; Frants, R.R.; Williamson, R.; Hewitt, J.E. Fine mapping of the FSHD gene region orientates the rearranged fragment detected by the probe p13E-11. Hum. Mol. Genet. 1993, 2, 1673–1678. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Papanikos, F.; Skoulatou, C.; Sakellariou, P.; Kekou, K.; Christopoulos, T.K.; Kanavakis, E.; Traeger-Synodinos, J.; Ioannou, P.C. A simplified approach for FSHD molecular testing. Clin. Chim. Acta 2014, 429, 96–103. [Google Scholar] [CrossRef]
- Van Overveld, P.G.M.; Lemmers, R.J.F.L.; Sandkuijl, L.A.; Enthoven, L.; Winokur, S.T.; Bakels, F.; Padberg, G.W.; Van Ommen, G.-J.B.; Frants, R.R.; van der Maarel, S. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003, 35, 315–317. [Google Scholar] [CrossRef]
- de Greef, J.; Lemmers, R.J.L.F.; Van Engelen, B.G.M.; Sacconi, S.; Venance, S.L.; Frants, R.R.; Tawil, R.; Van Der Maarel, S.M. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009, 30, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Dumbovic, G.; Forcales, S.-V.; Perucho, M. Emerging roles of macrosatellite repeats in genome organization and disease development. Epigenetics 2017, 12, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Shao, W.; Chang, L.; Yin, Y.; Li, T.; Zhang, H.; Hong, Y.; Percharde, M.; Guo, L.; Wu, Z.; et al. Genomic Repeats Categorize Genes with Distinct Functions for Orchestrated Regulation. Cell Rep. 2020, 30, 3296–3311.e5. [Google Scholar] [CrossRef] [PubMed]
- Lemmers, R.J.; Wohlgemuth, M.; van der Gaag, K.J.; van der Vliet, P.J.; van Teijlingen, C.M.; de Knijff, P.; Padberg, G.W.; Frants, R.R.; van der Maarel, S.M. Specific Sequence Variations within the 4q35 Region Are Associated with Facioscapulohumeral Muscular Dystrophy. Am. J. Hum. Genet. 2007, 81, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Scionti, I.; Greco, F.; Ricci, G.; Govi, M.; Arashiro, P.; Vercelli, L.; Berardinelli, A.; Angelini, C.; Antonini, G.; Cao, M.; et al. Large-Scale Population Analysis Challenges the Current Criteria for the Molecular Diagnosis of Fascioscapulohumeral Muscular Dystrophy. Am. J. Hum. Genet. 2012, 90, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.I.; Yan, C.; Sapp, P.C.; McKenna-Yasek, D.; Kang, P.B.; Quinn, C.; Salameh, J.S.; King, O.D.; Jones, P.L. Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin. Epigenetics 2014, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ricci, G.; Scionti, I.; Sera, F.; Govi, M.; D’Amico, R.; Frambolli, I.; Mele, F.; Filosto, M.; Vercelli, L.; Ruggiero, L.; et al. Large scale genotype–phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain 2013, 136, 3408–3417. [Google Scholar] [CrossRef]
- Zernov, N.; Skoblov, M. Genotype-phenotype correlations in FSHD. BMC Med. Genom. 2019, 12, 43. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Mah, J.K. A Pediatric Review of Facioscapulohumeral Muscular Dystrophy. J. Pediatr. Neurol. 2018, 16, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Rudnik-Schöneborn, S.; Huemer, M.; Weis, J.; Sauer, E.; Meng, G. Early onset facioscapulohumeral muscular dystrophy—Long-term follow-up of a patient with total facial diplegia. Neuromuscul. Disord. 2019, 29, 973–976. [Google Scholar] [CrossRef]
- Goselink, R.J.; Mul, K.; Van Kernebeek, C.R.; Lemmers, R.J.; van der Maarel, S.; Schreuder, T.H.; Erasmus, C.E.; Padberg, G.W.; Statland, J.M.; Voermans, N.C.; et al. Early onset as a marker for disease severity in facioscapulohumeral muscular dystrophy. Neurology 2019, 92, e378–e385. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zernov, N.V.; Guskova, A.A.; Skoblov, M.Y. FSHD1 Diagnosis in a Russian Population Using a qPCR-Based Approach. Diagnostics 2021, 11, 982. https://doi.org/10.3390/diagnostics11060982
Zernov NV, Guskova AA, Skoblov MY. FSHD1 Diagnosis in a Russian Population Using a qPCR-Based Approach. Diagnostics. 2021; 11(6):982. https://doi.org/10.3390/diagnostics11060982
Chicago/Turabian StyleZernov, Nikolay Vladimirovich, Anna Alekseevna Guskova, and Mikhail Yurevich Skoblov. 2021. "FSHD1 Diagnosis in a Russian Population Using a qPCR-Based Approach" Diagnostics 11, no. 6: 982. https://doi.org/10.3390/diagnostics11060982
APA StyleZernov, N. V., Guskova, A. A., & Skoblov, M. Y. (2021). FSHD1 Diagnosis in a Russian Population Using a qPCR-Based Approach. Diagnostics, 11(6), 982. https://doi.org/10.3390/diagnostics11060982





