MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Procedures
2.2. RNA Isolation
2.3. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
2.4. Statistical Analysis
3. Results
3.1. Basic Characteristics of the Study Subjects
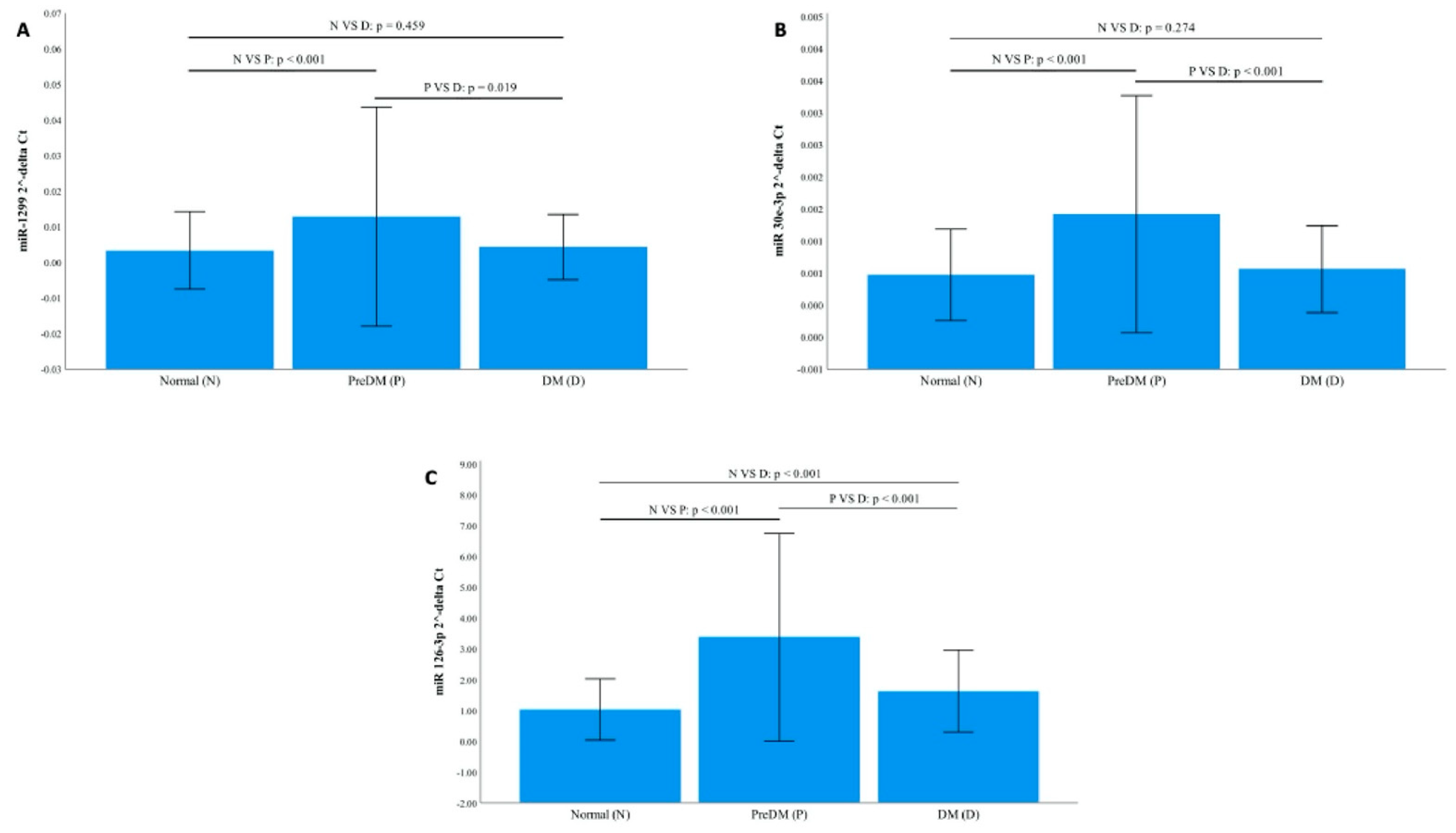
3.2. Relative Expression of microRNAs (miRNAs)
3.3. Correlation of miRNAs and Biochemical Parameters
3.4. Association between miRNAs and Prediabetes or Type 2 Diabetes
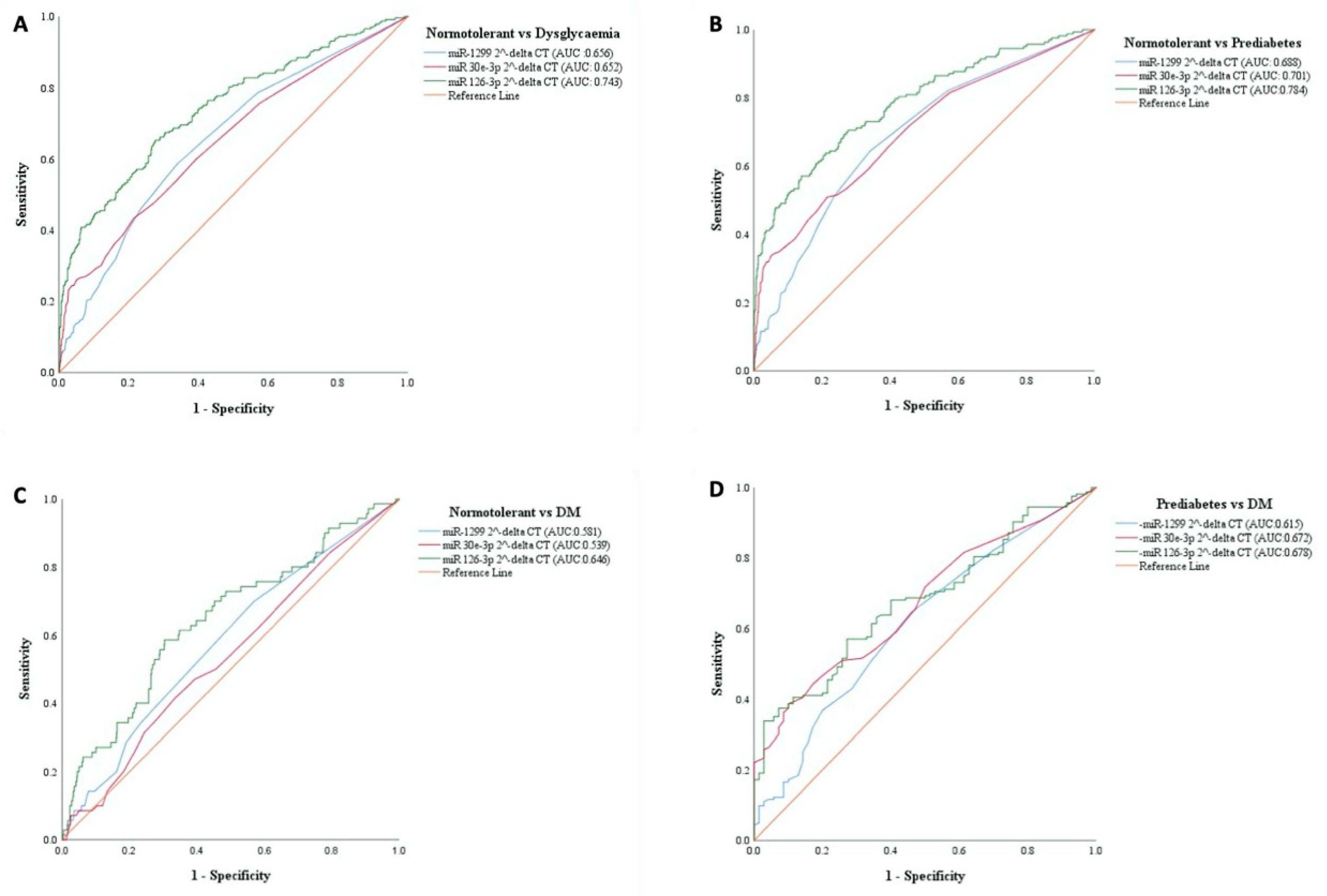
3.5. Diagnostic Specificity and Sensitivity of the miRNAs for Prediabetes and Type 2 Diabetes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Wang, G.; Liang, Y.; Zhou, X. Expression profiling and clinical significance of plasma microRNAs in diabetic nephropathy. J. Diabetes Res. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- de Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019; Available online: https://www.diabetesatlas.org (accessed on 8 April 2020).
- Goldenberg, R.; Punthakee, Z. Definition, classification and diagnosis of diabetes, prediabetes and metabolic syndrome. Can. J. Diabetes 2013, 37, S8–S11. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA profiling reveals loss of endothelial MiR-126 and other MicroRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, G.; Yang, C.; Zhou, K.; Shen, B.; Liang, H.; Jiang, X. The role of circulating microRNA-126 (miR-126): A novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int. J. Mol. Sci. 2014, 15, 10567–10577. [Google Scholar] [CrossRef] [PubMed]
- Amr, K.S.; Abdelmawgoud, H.; Ali, Z.Y.; Shehata, S.; Raslan, H.M. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br. J. Biomed. Sci. 2018, 75, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Matsha, T.E.; Kengne, A.P.; Hector, S.; Mbu, D.L.; Yako, Y.; Erasmus, R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget 2018, 9, 30485–30498. [Google Scholar] [CrossRef][Green Version]
- Kengne, A.P.; Erasmus, R.T.; Levitt, N.S.; Matsha, T.E. Alternative indices of glucose homeostasis as biochemical diagnostic tests for abnormal glucose tolerance in an African setting. Prim. Care Diabetes 2017, 11, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Stats, S.A. Statistics South Africa. In Formal Census 2011; Statistics South Africa: Pretoria, South Africa, 2011. [Google Scholar]
- Alberti, K.G.M.M.; Zimmet, P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus. Provisional report of a WHO consultation. Diabet. Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Weale, C.J.; Matshazi, D.M.; Davids, S.F.G.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. Circulating miR-30a-5p and miR-182-5p in Prediabetes and Screen-Detected Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2020, 13, 5037–5047. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Tang, G.; Özcan, S. Role of MicroRNAs in Diabetes. Biochim. Biophys. Acta 2008, 1779, 697–701. [Google Scholar] [CrossRef]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.G. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, Z.; Wang, Z.; Zeng, F.; Xiao, W.; Yu, A. MicroRNA-126: A promising biomarker for angiogenesis of diabetic wounds treated with negative pressure wound therapy. Diabetes Metab. Syndr. Obes. 2019, 12, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Sen, C.K. miRNA in wound inflammation and angiogenesis. Microcirculation 2012, 19, 224–232. [Google Scholar] [CrossRef]
- Zhang, T.; Lv, C.; Li, L.; Chen, S.; Liu, S.; Wang, C.; Su, B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed. Res. Int. 2013, 2013, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaji, G.; Al-Mahroos, G.; Al-Muhtaresh, H.A.; Skrypnyk, C.; Sabry, M.A.; Ramadan, A.R. Decreased expression of circulating microRNA-126 in patients with type 2 diabetic nephropathy: A potential blood-based biomarker. Exp. Ther. Med. 2016, 12, 815–822. [Google Scholar] [CrossRef]
- Barutta, F.; Bellini, S.; Mastrocola, R.; Bruno, G.; Gruden, G. MicroRNA and Microvascular Complications of Diabetes. Int. J. Endocrinol. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.-L.; An, M.-X.; Liu, Y.-L.; Xu, H.-C.; Lu, Z.-Q. MicroRNA-126: A promising novel biomarker in peripheral blood for diabetic retinopathy. Int. J. Ophthalmol. 2017, 10, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Rawal, S.; Munasinghe, P.E.; Shindikar, A.; Paulin, J.; Cameron, V.; Manning, P.; Williams, M.J.; Jones, G.T.; Bunton, R.; Galvin, I.; et al. Down-regulation of proangiogenic microRNA-126 and microRNA-132 are early modulators of diabetic cardiac microangiopathy. Cardiovasc. Res. 2017, 113, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Rezk, N.A.; Sabbah, N.A.; Saad, M.S.S. Role of microRNA 126 in screening, diagnosis, and prognosis of diabetic patients in Egypt. IUBMB Life 2016, 68, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Papaneophytou, C. Challenges in Using Circulating Micro-RNAs as Biomarkers for Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.-H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef]
- Zhang, F.B.; Du, Y.; Tian, Y.; Ji, Z.G.; Yang, P.Q. MiR-1299 functions as a tumor suppressor to inhibit the proliferation and metastasis of prostate cancer by targeting NEK2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 530–538. [Google Scholar] [PubMed]
- Zhu, H.; Wang, G.; Zhou, X.; Song, X.; Gao, H.; Ma, C.; Chang, H.; Li, H.; Liu, F.F.; Lu, J.; et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed. Pharmacother. 2016, 83, 792–797. [Google Scholar] [CrossRef]
- Li, N.; Lian, J.; Zhao, S.; Zheng, D.; Yang, X.; Huang, X.; Shi, X.; Sun, L.; Zhou, Q.; Shi, H.; et al. Detection of Differentially Expressed MicroRNAs in Rheumatic Heart Disease: MiR-1183 and miR-1299 as Potential Diagnostic Biomarkers. Biomed. Res. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Massaro, J.D.; Polli, C.D.; e Silva, M.C.; Alves, C.C.; Passos, G.A.; Sakamoto-Hojo, E.T.; de Holanda Miranda, W.R.; Cezar, N.J.B.; Rassi, D.M.; Crispim, F.; et al. Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol. Cell. Endocrinol. 2019, 490, 1–14. [Google Scholar] [CrossRef]
- Liu, M.M.; Li, Z.; Han, X.D.; Shi, J.H.; Tu, D.Y.; Song, W.; Zhang, J.; Qiu, X.L.; Ren, Y.; Zhen, L.L. MiR-30e inhibits tumor growth and chemoresistance via targeting IRS1 in Breast Cancer. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Steele, R.; Shrivastava, S.; Chakraborty, S.; Di Bisceglie, A.M.; Ray, R.B. Serum miR-30e and miR-223 as novel noninvasive biomarkers for hepatocellular Carcinoma. Am. J. Pathol. 2016, 186, 242–247. [Google Scholar] [CrossRef] [PubMed]
| Variable | NGT, n = 972 | Prediabetes, n = 207 | DM, n = 94 | p-Value |
|---|---|---|---|---|
| Age (years) | 45.2 ± 15.3 | 55.1 ± 13 | 58.4 ± 10.6 | <0.001 |
| Male, n (%) | 284 (29.3) | 42 (20.3) | 19 (20.2) | |
| Body mass index (kg/m2) | 27.4 ± 7.9 | 31.2 ± 8.7 | 31.3 ± 8 | <0.001 |
| Waist circumference (cm) | 88.1 ± 16.8 | 97 ± 15.8 | 99.9 ± 15.5 | <0.001 |
| Hip circumference (cm) | 101.1 ± 16.7 | 107.7 ± 16.5 | 107.8 ± 15.3 | <0.001 |
| Waist to hip ratio | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | <0.001 |
| Systolic blood pressure (mmHg) | 131 ± 25 | 145 ± 27.3 | 145.2 ± 25.5 | <0.001 |
| Diastolic blood pressure (mmHg) | 83.6 ± 15.1 | 89.9 ± 15.4 | 89.6 ± 13.6 | <0.001 |
| Fasting glucose (mmol/L | 4.7 ± 0.5 | 5.4 ± 0.7 | 8.3 ± 3.9 | <0.001 |
| Post 2 h glucose (mmol/L) * | 5.4 (4.5; 6.3) | 8.6 (8; 9.6) | 12.9 (11.6; 16.8) | <0.001 |
| HbA1c (%) | 5.6 ± 0.5 | 5.9 ± 0.5 | 7.4 ± 2.1 | <0.001 |
| HbA1c (mmol/mol) | 37.7 | 41.0 | 57.4 | |
| Fasting insulin (mIU/L) | 7.6 ± 7.2 | 11.3 ± 11.3 | 14.4 ± 29.5 | <0.001 |
| Post 2-h insulin (mIU/L) * | 30.5 (16; 53.6) | 71.6 (42.3; 113.2) | 50.5 (29.1; 79.4) | <0.001 |
| Triglycerides (mmol/L) * | 1.1 (0.8; 1.5) | 1.4 (1; 1.8) | 1.4 (1.1; 2.4) | <0.001 |
| HDL-cholesterol (mmol/L) | 1.4 ± 0.4 | 1.4 ± 0.4 | 1.3 ± 0.5 | 0.640 |
| LDL-cholesterol (mmol/L) | 3.1 ± 1 | 3.3 ± 0.9 | 3.5 ± 1.1 | <0.001 |
| usCRP (mg/L) | 3.4 (1.3; 7.7) | 5 (2.2; 11.0) | 6.5 (3.3; 13.1) | <0.001 |
| Cotinine (ng/mL) * | 120.5 (10; 285.3) | 10 (10; 271.5) | 10 (10; 183) | <0.001 |
| GGT (IU/L) | 27 (19; 42) | 31 (22; 53) | 42 (25.5; 76) | <0.001 |
| Current smokers, n(%) | 540 (57.8) | 99 (48.3) | 29 (32.6) | <0.001 |
| Current drinker, n(%) | 322 (33.3) | 56 (27.5) | 15 (16.1) | 0.002 |
| MicroRNA | Prediabetes vs. NGT | DM vs. NGT | DM vs. Prediabetes |
|---|---|---|---|
| RT-qPCR | |||
| miR-1299 | 4.17 ± 0.10 | 1.99 ± 0.13 | 0.48 ± 0.06 |
| miR-30e-3p | 3.22 ± 0.07 | 1.32 ± 0.17 | 0.41 ± 0.13 |
| miR-126-3p | 3.12 ± 0.11 | 1.75 ± 0.03 | 0.56 ± 0.09 |
| NGS Fold Changes [9] * | |||
| miR-1299 | 5.38 ± 0.23 | 1 ± 0.90 | 0.72 ± 0.88 |
| miR-30e-3p | 2.40 ± 0.03 | 1.78 ± 0.1 | 0.51 ± 1.15 |
| miR-126-3p | 1.74 ± 0.10 | 1 ± 1.2 | 1.53 ± 0.05 |
| Variable | miR-1299 | miR-30e-3p | miR-126-3p | |||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| miR-1299 | 1.000 | 0.712 | 0.000 | 0.731 | <0.001 | |
| miR-30e-3p | 0.712 | <0.001 | 1.000 | 0.965 | <0.001 | |
| miR-126-3p | 0.731 | <0.001 | 0.965 | <0.001 | 1.000 | |
| Waist circumference (cm) | −0.465 | 0.039 | −0.471 | 0.036 | −0.444 | 0.050 |
| Hip circumference (cm) | 0.150 | 0.527 | 0.027 | 0.909 | 0.053 | 0.823 |
| Waist hip ratio | −0.113 | 0.635 | −0.076 | 0.751 | −0.083 | 0.727 |
| Systolic blood pressure (mmHg) | 0.198 | 0.403 | 0.197 | 0.406 | 0.193 | 0.415 |
| Diastolic blood pressure (mmHg) | 0.201 | 0.395 | 0.171 | 0.472 | 0.161 | 0.497 |
| Fasting glucose (mmol/L) | 0.176 | 0.457 | 0.264 | 0.261 | 0.253 | 0.281 |
| Post 2-h glucose (mmol/L) | 0.369 | 0.109 | 0.399 | 0.082 | 0.429 | 0.059 |
| HbA1c (mmol/mol) | 0.072 | 0.764 | 0.055 | 0.819 | 0.061 | 0.799 |
| Fasting insulin (mIU/L) | 0.265 | 0.258 | 0.292 | 0.211 | 0.307 | 0.187 |
| Post 2-h insulin (mIU/L) | 0.202 | 0.392 | 0.214 | 0.364 | 0.241 | 0.306 |
| Triglycerides-S (mmol/L) | −0.049 | 0.839 | 0.015 | 0.949 | 0.071 | 0.765 |
| HDL-cholesterol (mmol/L) | 0.458 | 0.042 | 0.452 | 0.045 | 0.401 | 0.079 |
| LDL-cholesterol (mmol/L) | 0.070 | 0.771 | 0.031 | 0.895 | 0.047 | 0.845 |
| usCRP (mg/L) | 0.185 | 0.435 | 0.199 | 0.400 | 0.165 | 0.488 |
| Cotinine (ng/mL) | 0.434 | 0.056 | 0.393 | 0.086 | 0.381 | 0.098 |
| GGT (IU/L) | 0.117 | 0.623 | 0.125 | 0.598 | 0.121 | 0.610 |
| MicroRNA | Prediabetes | DM | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p-Value | OR | 95% CI | p-Value | |
| miR-1299 * | ||||||
| Model 1 | 1.37 | (1.20; 1.55) | <0.001 | 1.12 | (0.89; 1.39) | 0.338 |
| Model 2 | 1.37 | (1.20; 1.56) | <0.001 | 1.11 | (0.89; 1.40) | 0.354 |
| Model 3 | 1.41 | (1.22; 1.61) | <0.001 | 1.14 | (0.91; 1.44) | 0.254 |
| Model 4 | 1.41 | (1.21; 1.64) | <0.001 | 1.17 | (0.92; 1.49) | 0.213 |
| Model 5 | 1.26 | (1.05; 1.50) | 0.012 | 0.92 | (0.56; 1.51) | 0.744 |
| Model 6 | 1.31 | (1.07; 1.60) | 0.009 | 0.87 | (0.48; 1.56) | 0.634 |
| miR-30e-3p * | ||||||
| Model 1 | 2.10 | (1.78; 2.48) | <0.001 | 1.19 | (0.89; 1.59) | 0.241 |
| Model 2 | 2.17 | (1.83; 2.58) | <0.001 | 1.26 | (0.94; 1.69) | 0.117 |
| Model 3 | 2.16 | (1.82; 2.57) | <0.001 | 1.26 | (0.94; 1.69) | 0.127 |
| Model 4 | 2.11 | (1.77; 2.51) | <0.001 | 1.20 | (0.88; 1.65) | 0.247 |
| Model 5 | 1.94 | (1.37; 2.74) | <0.001 | 1.29 | (0.69; 2.41) | 0.433 |
| Model 6 | 2.04 | (1.36; 3.08) | 0.001 | 1.32 | (0.68; 2.59) | 0.414 |
| miR-126-3p ** | ||||||
| Model 1 | 2.07 | (1.85; 2.33) | <0.001 | 1.46 | (1.25; 1.70) | <0.001 |
| Model 2 | 2.15 | (1.90; 2.43) | <0.001 | 1.53 | (1.31; 1.80) | <0.001 |
| Model 3 | 2.13 | (1.88; 2.41) | <0.001 | 1.52 | (1.29; 1.78) | <0.001 |
| Model 4 | 2.05 | (1.81; 2.33) | <0.001 | 1.43 | (1.21; 1.69) | <0.001 |
| Model 5 | 1.91 | (1.51; 2.41) | <0.001 | 1.65 | (1.19; 2.30) | <0.001 |
| Model 6 | 2.04 | (1.56; 2.68) | <0.001 | 1.80 | (1.25; 2.60) | <0.001 |
| MicroRNA | Prediabetes | DM | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p-Value | OR | 95% CI | p-Value | |
| miR-1299 * | ||||||
| Model 1 | - | - | - | 0.82 | (0.66; 1.01) | 0.065 |
| Model 2 | - | - | - | 0.81 | (0.65; 1.01) | 0.068 |
| Model 3 | - | - | - | 0.81 | (0.65; 1.02) | 0.068 |
| Model 4 | - | - | - | 0.83 | (0.66; 1.04) | 0.098 |
| Model 5 | - | - | - | 0.73 | (0.46; 1.16) | 0.188 |
| Model 6 | - | - | - | 0.66 | (0.38; 1.15) | 0.145 |
| miR-30e-3p * | ||||||
| Model 1 | - | - | - | 0.57 | (0.42; 0.76) | <0.001 |
| Model 2 | - | - | - | 0.58 | (0.43; 0.78) | <0.001 |
| Model 3 | - | - | - | 0.58 | (0.43; 0.78) | <0.001 |
| Model 4 | - | - | - | 0.57 | (0.42; 0.78) | <0.001 |
| Model 5 | - | - | - | 0.67 | (0.40; 1.14) | 0.138 |
| Model 6 | - | - | - | 0.65 | (0.38; 1.11) | 0.117 |
| miR-126-3p ** | ||||||
| Model 1 | - | - | - | 0.70 | (0.61; 0.82) | <0.001 |
| Model 2 | - | - | - | 0.71 | (0.62; 0.83) | <0.001 |
| Model 3 | - | - | - | 0.71 | (0.61; 0.83) | <0.001 |
| Model 4 | - | - | - | 0.70 | (0.59; 0.81) | <0.001 |
| Model 5 | - | - | - | 0.87 | (0.69; 1.10) | 0.235 |
| Model 6 | - | - | - | 0.88 | (0.69; 1.13) | 0.328 |
| Performance Measure | Dysglycemia | Prediabetes | Diabetes | |||
|---|---|---|---|---|---|---|
| miR-126-3p | HbA1c | miR-126-3p | HbA1c | miR-126-3p | HbA1c | |
| AUC | 0.743 (0.705–0.781) | 0.753 (0.717–0.788) | 0.784 (0.742–0.827) | 0.695 (0.652–0.739) | 0.646 (0.576–0.717) | 0.861 (0.812–0.909) |
| Threshold | 1.41 (1.25–1.84) | 5.95 (5.75–6.05) | 1.78 (1.38–3.07) | 5.75 (5.75–9.95) | 1.31 (0.60–1.42) | 6.05 (6.05–6.45) |
| Sensitivity | 0.628 (0.570–0.683) | 0.591 (0.497–0.733) | 0.616 (0.545–0.683) | 0.598 (0.461–0.721) | 0.556 (0.447–0.660) | 0.761 (0.611–0.859) |
| Specificity | 0.737 (0.708–0.765) | 0.824 (0.670–0.893) | 0.804 (0.777–0.828) | 0.707 (0.643–0.846) | 0.708 (0.678–0.737) | 0.837 (0.807–0.959) |
| PPV | 0.424 (0.391–0.458) | 0.519 (0.410–0.616) | 0.401 (0.361–0.442) | 0.335 (0.234–0.430) | 0.152 (0.127–0.182) | 0.284 (0.239–0.566) |
| NPV | 0.866 (0.847–0.882) | 0.865 (0.846–0.891) | 0.907 (0.892–0.921) | 0.894 (0.875–0.916) | 0.944 (0.930–0.955) | 0.978 (0.968–0.986) |
| Accuracy | 0.718 (0.686–0.737) | 0.771 (0.685–0.810) | 0.760 (0.734–0.784) | 0.702 (0.647–0.789) | 0.684 (0.655–0.711) | 0.833 (0.805–0.938) |
| p-value for AUC Comparison | 0.281 | 0.048 | <0.001 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weale, C.J.; Matshazi, D.M.; Davids, S.F.G.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes. Diagnostics 2021, 11, 949. https://doi.org/10.3390/diagnostics11060949
Weale CJ, Matshazi DM, Davids SFG, Raghubeer S, Erasmus RT, Kengne AP, Davison GM, Matsha TE. MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes. Diagnostics. 2021; 11(6):949. https://doi.org/10.3390/diagnostics11060949
Chicago/Turabian StyleWeale, Cecil J., Don M. Matshazi, Saarah F. G. Davids, Shanel Raghubeer, Rajiv T. Erasmus, Andre P. Kengne, Glenda M. Davison, and Tandi E. Matsha. 2021. "MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes" Diagnostics 11, no. 6: 949. https://doi.org/10.3390/diagnostics11060949
APA StyleWeale, C. J., Matshazi, D. M., Davids, S. F. G., Raghubeer, S., Erasmus, R. T., Kengne, A. P., Davison, G. M., & Matsha, T. E. (2021). MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes. Diagnostics, 11(6), 949. https://doi.org/10.3390/diagnostics11060949







