Multicentre Evaluation of Hepika Test Clinical Accuracy in Diagnosing HPV-Induced Cancer and Precancerous Lesions of the Uterine Cervix
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Definition of the Reference Standard
2.3. Evaluation of Sample Adequacy and Cytology
- (a)
- Total number of optical fields 20× (FOV20×) in a slide = µm2 314,000,000 of circular area of slide containing the cells/µm2 350,000 of area of FOV20× = 897.
- (b1)
- Number of total cells estimated for FOV20× = Mean of the count of total cells found in 20 FOV20× of slide.
- (b2)
- Number of atypical cells estimated for FOV20× = Mean of the count of atypical cells found in 20 FOV20× of slide.
- (c1)
- Total cells present in a slide = (a × b1) = (897 × b1).
- (c2)
- Atypical cells present in a slide = (a × b2) = (897 × b2).
- (d1)
- Total cells present in residual sample = (c1/5 mL × n mL of residual liquid).
- (d2)
- Atypical cells present in residual sample = (c2/5 mL × n mL of residual liquid).
2.4. Hepika® Test (CE-IVD)
2.5. Human Papillomavirus (HPV) DNA Test
2.6. Statistical Analysis
3. Results
Adequacy of the Sample
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | Adenocarcinoma |
| AGC | Atypical glandular cells |
| AIS | Adenocarcinoma in situ |
| ASC-H | Atypical squamous cells–cannot exclude HSIL |
| ASC-US | Atypical squamous cells of undetermined significance |
| CC | Carcinoma |
| CIN | Cervical intraepithelial neoplasia |
| CIN1 | Cervical intraepithelial neoplasia grade 1 |
| CIN2 | Cervical intraepithelial neoplasia grade 2 |
| CIN2+ | Cervical intraepithelial neoplasia grade 2 or worse |
| CIN3 | Cervical intraepithelial neoplasia grade 3 |
| CIN3+ | Cervical intraepithelial neoplasia grade 3 or worse |
| HG | High grade |
| HK | Hepika |
| HK+ | Hepika-positive |
| HK− | Hepika-negative |
| HPV+ | HPV-positive |
| HPV− | HPV-negative |
| HSIL | High-grade squamous intraepithelial lesion |
| LG | Low grade |
| LSIL | Low-grade squamous intraepithelial lesion |
| LR+ | Positive likelihood ratio |
| LR− | Negative likelihood ratio |
| n | Number of cases |
| N.P. | Not performed |
| NEG | Negative |
| NPV | Negative predictive value |
| POS | Positive |
| PPV | Positive predictive value |
| SCC | Squamous cell carcinoma |
References
- WHO. Human Papillomavirus (HPV) and Cervical Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/human-papillomavirus-(hpv)-and-cervical-cancer (accessed on 30 January 2019).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of inci-dence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Vizcaino, A.P.; Moreno, V.; Bosch, F.X.; Muñoz, N.; Barros-Dios, X.M.; Parkin, D.M. International trends in the incidence of cervical cancer: I. Adenocarcinoma and adenosquamous cell carcinomas. Int. J. Cancer 1998, 75, 536–545. [Google Scholar] [CrossRef]
- Gustafsson, L.; Pontén, J.; Zack, M.; Adami, H.-O. International incidence rates of invasive cervical cancer after introduction of cytological screening. Cancer Causes Control. 1997, 8, 755–763. [Google Scholar] [CrossRef]
- Gustafsson, L.; Pontén, J.; Bergstrôm, R.; Adami, H.-O. International incidence rates of invasive cervical cancer before cytological screening. Int. J. Cancer 1997, 71, 159–165. [Google Scholar] [CrossRef]
- IARC. Working Group on Cervical Cancer Screening and the UICC Project Group on the Evaluation of Screening Programmes for Cancer. Screening for cancer of the uterine cervix. IARC Sci. Publ. 1986, 76, 1–315. [Google Scholar]
- Lorincz, A.; Castanon, A.; Lim, A.W.W.; Sasieni, P. New Strategies for Papillomavirus-Based Cervical Screening. Women’s Health 2013, 9, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Bosch, F.X.; Lorincz, A.; Munoz, N.; Meijer, C.J.L.M.; Shah, K.V. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef] [PubMed]
- Dillner, J.; Rebolj, M.; Birembaut, P.; Petry, K.-U.; Szarewski, A.; Munk, C.; De Sanjose, S.; Naucler, P.; Lloveras, B.; Kjaer, S.; et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: Joint European cohort study. BMJ 2008, 13, 337. [Google Scholar] [CrossRef] [PubMed]
- Ronco, G.; Dillner, J.; Elfström, K.M.; Tunesi, S.; Snijders, P.J.F.; Arbyn, M.; Kitchener, H.; Segnan, N.; Gilham, C.; Giorgi-Rossi, P.; et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: Follow-up of four European randomised controlled trials. Lancet 2014, 383, 524–532. [Google Scholar] [CrossRef]
- US Preventive Services Task Force. Screening for cervical cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018, 320, 674–686. [Google Scholar] [CrossRef]
- Anttila, A.; Arbyn, M.; Bergeron, C.; Cuzick, J.; De Vuyst, H.; Dillner, J.; Dillner, L.; Ducarroz, S.; Franceschi, S.; Giorgi Rossi, P.; et al. European Guidelines for Quality Assurance in Cervical Cancer Screening—Second Edition. Available online: https://op.europa.eu/en/publication-detail/-/publication/a41a4c40-0626-4556-af5b-2619dd1d5ddc/language-en (accessed on 20 April 2018).
- Ministero Della Salute. Piano Nazionale Della Prevenzione 2014–2018. Available online: http://www.salute.gov.it/imgs/C_17_pubblicazioni_2285_allegato.pdf (accessed on 24 March 2020).
- Ronco, G.; Giorgi-Rossi, P.; Carozzi, F.; Confortini, M.; Palma, P.D.; Del Mistro, A.; Gillio-Tos, A.; Minucci, D.; Naldoni, C.; Rizzolo, R.; et al. Results at Recruitment From a Randomized Controlled Trial Comparing Human Papillomavirus Testing Alone With Conventional Cytology as the Primary Cervical Cancer Screening Test. J. Natl. Cancer Inst. 2008, 100, 492–501. [Google Scholar] [CrossRef]
- Ronco, G.; Giorgi-Rossi, P.; Carozzi, F.; Confortini, M.; Palma, P.D.; Del Mistro, A.; Ghiringhello, B.; Girlando, S.; Gillio-Tos, A.; De Marco, L.; et al. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: A randomised controlled trial. Lancet Oncol. 2010, 11, 249–257. [Google Scholar] [CrossRef]
- zur Hausen, H. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin. Cancer Biol. 1999, 9, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Hausen, H.Z. Papillomaviruses Causing Cancer: Evasion From Host-Cell Control in Early Events in Carcinogenesis. J. Natl. Cancer Inst. 2000, 92, 690–698. [Google Scholar] [CrossRef] [PubMed]
- McCredie, M.R.; Sharples, K.J.; Paul, C.; Baranyai, J.; Medley, G.; Jones, R.W.; Skegg, D.C. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: A retrospective cohort study. Lancet Oncol. 2008, 9, 425–434. [Google Scholar] [CrossRef]
- van den Heuvel, S. Cell-cycle regulation. WormBook 2005, 1–16. [Google Scholar] [CrossRef]
- Fehrmann, F.; A Laimins, L. Human papillomaviruses: Targeting differentiating epithelial cells for malignant transformation. Oncogene 2003, 22, 5201–5207. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef]
- Buitrago-Pérez, A.; Garaulet, G.; Vázquez-Carballo, A.; Paramio, J.M.; García-Escudero, R. Molecular Signature of HPV-Induced Carcinogenesis: pRb, p53 and Gene Expression Profiling. Curr. Genom. 2009, 10, 26–34. [Google Scholar] [CrossRef]
- Ishiji, T. Molecular Mechanism of Carcinogenesis by Human Papillomavirus-16. J. Dermatol. 2000, 27, 73–86. [Google Scholar] [CrossRef]
- Shaikh, F.; Sanehi, P.; Rawal, R. Molecular screening of compounds to the predicted Protein-Protein Interaction site of Rb1-E7 with p53-E6 in HPV. Bioinformation 2012, 8, 607–612. [Google Scholar] [CrossRef]
- de Sanjose, S.; Quint, W.G.; Alemany, L.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R.; Vallejos, C.S.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Thomas, M.; Pim, D.; Banks, L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999, 18, 7690–7700. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Pim, D.; Banks, L. Interaction of viral oncoproteins with cellular target molecules: Infection with high-risk vs low-risk human papillomaviruses. APMIS 2010, 118, 471–493. [Google Scholar] [CrossRef] [PubMed]
- Mammas, I.N.; Sourvinos, G.; Giannoudis, A.; Spandidos, D.A. Human Papilloma Virus (HPV) and Host Cellular Interactions. Pathol. Oncol. Res. 2008, 14, 345–354. [Google Scholar] [CrossRef]
- DeFilippis, R.A.; Goodwin, E.C.; Wu, L.; DiMaio, D. Endogenous Human Papillomavirus E6 and E7 Proteins Differentially Regulate Proliferation, Senescence, and Apoptosis in HeLa Cervical Carcinoma Cells. J. Virol. 2003, 77, 1551–1563. [Google Scholar] [CrossRef]
- Kang, H.T.; Lee, C.J.; Seo, E.J.; Bahn, Y.J.; Kim, H.J.; Hwang, E.S. Transition to an irreversible state of senescence in HeLa cells arrested by repression of HPV E6 and E7 genes. Mech. Ageing Dev. 2004, 125, 31–40. [Google Scholar] [CrossRef]
- Di Benedetto, G.; Maccallini, V. The Detection of Complexed Proteins E6/P53 and E7/Prb In Relation to the Carcinogenesis of the Uterine Cervix. J. Cytol. Histol. 2014, 2, S4. [Google Scholar] [CrossRef]
- Smeets, S.J.; Van Der Plas, M.; Schaaij-Visser, T.B.; Van Veen, E.A.; Van Meerloo, J.; Braakhuis, B.J.; Steenbergen, R.D.; Brakenhoff, R.H. Immortalization of oral keratinocytes by functional inactivation of the p53 and pRb pathways. Int. J. Cancer 2011, 128, 1596–1605. [Google Scholar] [CrossRef]
- GISCi. Gruppo Italiano Screening Cervicale. Raccomandazioni sul Test HR-HPV Come Test di Screening Primario e Rivisitazione del Ruolo del Pap Test. 2010. Available online: https://www.gisci.it/documenti/documenti_gisci/documento_hpv.pdf (accessed on 24 March 2021).
- Nayar, R.; Solomon, D. The Bethesda System for Reporting Cervical Cytology; Springer: New York, NY, USA, 2004. [Google Scholar]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. WHO Classification of Tumours of Female Reproductive Organs; IARC: Lyon, France, 2014. [Google Scholar]
- Grapsa, D.; Ioakim-Liossi, A.; Stergiou, E.; Petrakakou, E.; Nicolopoulou-Stamati, P.; Patsouris, E.; Athanassiadou, P. Repeat Processing of Residual ThinPrep Pap Tests: Sampling of the Vial May Not Be Invariably Homogeneous. Acta Cytol. 2011, 55, 213–217. [Google Scholar] [CrossRef]
- Stoler, M.H.; Wright, T.C., Jr.; Sharma, A.; Apple, R.; Gutekunst, K.; Wright, T.L.; ATHENA (Addressing THE Need for Advanced HPV Diagnostics) HPV Study Group. High-risk human papillomavirus testing in women with ASC-US cytology: Results from the ATHENA HPV study. Am. J. Clin. Pathol. 2011, 135, 468–475. [Google Scholar] [CrossRef]
- Clavel, C.; Masure, M.; Levert, M.; Putaud, I.; Mangeonjean, C.; Lorenzato, M.; Nazeyrollas, P.; Gabriel, R.; Quereux, C.; Birembaut, P. Human Papillomavirus Detection by the Hybrid Capture II Assay: A Reliable Test to Select Women With Normal Cervical Smears at Risk for Developing Cervical Lesions. Diagn. Mol. Pathol. 2000, 9, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, C.; Ronco, G.; Reuschenbach, M.; Wentzensen, N.; Arbyn, M.; Stoler, M.; von Knebel Doeberitz, M. The clinical impact of using p16INK4a immunochemistry in cervical histopathology and cytology: An update of recent developments. Int. J. Cancer 2015, 136, 2741–2751. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Clarke, M.A.; Bremer, R.; Poitras, N.; Tokugawa, D.; Goldhoff, P.E.; Castle, P.E.; Schiffman, M.; Kingery, J.D.; Grewal, K.K.; et al. Clinical Evaluation of Human Papillomavirus Screening with p16/Ki-67 Dual Stain Triage in a Large Organized Cervical Cancer Screening Program. JAMA Intern. Med. 2019, 179, 881–888. [Google Scholar] [CrossRef]
- Carozzi, F.; Ronco, G.; Gillio-Tos, A.; De Marco, L.; Del Mistro, A.; Girlando, S.; Franceschi, S.; Plummer, M.; Vaccarella, S.; New Technologies for Cervical Cancer screening (NTCC) Working Group. Concurrent infections with multiple human papillomavirus (HPV) types in the New Technologies for Cervical Cancer (NTCC) screening study. Eur. J. Cancer 2012, 48, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Gustinucci, D.; Rossi, P.G.; Cesarini, E.; Broccolini, M.; Bulletti, S.; Carlani, A.; D’Angelo, V.; D’Amico, M.R.; Di Dato, E.; Galeazzi, P.; et al. Use of Cytology, E6/E7 mRNA, and p16INK4a–Ki-67 to Define the Management of Human Papillomavirus (HPV)–Positive Women in Cervical Cancer Screening. Am. J. Clin. Pathol. 2016, 145, 35–45. [Google Scholar] [CrossRef]
- Carozzi, F.; Confortini, M.; Palma, P.D.; del Mistro, A.; Gillio-Tos, A.; de Marco, L.; Giorgi-Rossi, P.; Pontenani, G.; Rosso, S.; Sani, C.; et al. Use of p16-INK4A overexpression to increase the specificity of human papillomavirus testing: A nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2008, 9, 937–945. [Google Scholar] [CrossRef]
- Benevolo, M.; Vocaturo, A.; Mottolese, M.; Mariani, L.; Vocaturo, G.; Marandino, F.; Sperduti, I.; Rollo, F.; Antoniani, B.; Donnorso, R.P.; et al. Clinical role of p16INK4a expression in liquid-based cervical cytology: Correlation with HPV testing and histologic diagnosis. Am. J. Clin. Pathol. 2008, 129, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Carozzi, F.; Gillio-Tos, A.; Confortini, M.; Del Mistro, A.; Sani, C.; De Marco, L.; Girlando, S.; Rosso, S.; Naldoni, C.; Palma, P.D.; et al. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: A prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2013, 14, 168–176. [Google Scholar] [CrossRef]
- Pacchiarotti, A.; Ferrari, F.; Bellardini, P.; Chini, F.; Collina, G.; Palma, P.D.; Ghiringhello, B.; Maccallini, V.; Musolino, F.; Negri, G.; et al. Prognostic value of p16-INK4A protein in women with negative or CIN1 histology result: A follow-up study. Int. J. Cancer 2013, 134, 897–904. [Google Scholar] [CrossRef]
- Yang, Y.-S.; Smith-McCune, K.; Darragh, T.M.; Lai, Y.; Lin, J.-H.; Chang, T.-C.; Guo, H.-Y.; Kesler, T.; Carter, A.; Castle, P.E.; et al. Direct Human Papillomavirus E6 Whole-Cell Enzyme-Linked Immunosorbent Assay for Objective Measurement of E6 Oncoproteins in Cytology Samples. Clin. Vaccine Immunol. 2012, 19, 1474–1479. [Google Scholar] [CrossRef]
- Jeon, S.; Allen-Hoffmann, B.L.; Lambert, P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [CrossRef]
- Werness, B.A.; Levine, A.J.; Howley, P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990, 248, 76–79. [Google Scholar] [CrossRef]
- Ghittoni, R.; Accardi, R.; Hasan, U.; Gheit, T.; Sylla, B.; Tommasino, M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 2010, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2016, 25, 2–23. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Vinokurova, S.; Doeberitz, M.V.K. Systematic Review of Genomic Integration Sites of Human Papillomavirus Genomes in Epithelial Dysplasia and Invasive Cancer of the Female Lower Genital Tract. Cancer Res. 2004, 64, 3878–3884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shen, C.; Zhao, L.; Wang, J.; A McCrae, M.; Chen, X.; Lu, F. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV-related cervical carcinogenesis. Int. J. Cancer 2016, 138, 1163–1174. [Google Scholar] [CrossRef]





| Histology | HPV | Cytology | Total | % | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | % | NEG | % | ADC | SCC | HSIL | LSIL | ASC-H | ASC-US | AGC | NEG | N.P. | |||
| ADC | 4 | 1.5% | 2 | 1 | 1 | 4 | 1.5% | ||||||||
| SCC | 6 | 2.2% | 1 | 0.4% | 3 | 3 | 1 | 7 | 2.6% | ||||||
| AIS | 1 | 0.4% | 1 | 1 | 0.4% | ||||||||||
| CIN3 | 60 | 21.9% | 1 | 43 | 9 | 2 | 5 | 60 | 21.9% | ||||||
| CIN2 | 51 | 18.6% | 20 | 19 | 4 | 8 | 51 | 18.6% | |||||||
| CIN1 | 34 | 12.4% | 7 | 19 | 2 | 6 | 34 | 12.4% | |||||||
| NEG | 49 | 17.9% | 68 | 24.8% | 1 | 18 | 16 | 1 | 2 | 1 | 13 | 65 | 117 | 42.7% | |
| Total | 205 | 74.8% | 69 | 25.2% | 2 | 5 | 92 | 63 | 10 | 2 | 2 | 32 | 66 | 274 | 100.0% |
| 0.7% | 1.8% | 33.6% | 23.0% | 3.6% | 0.7% | 0.7% | 11.7% | 24.1% | 100.0% | ||||||
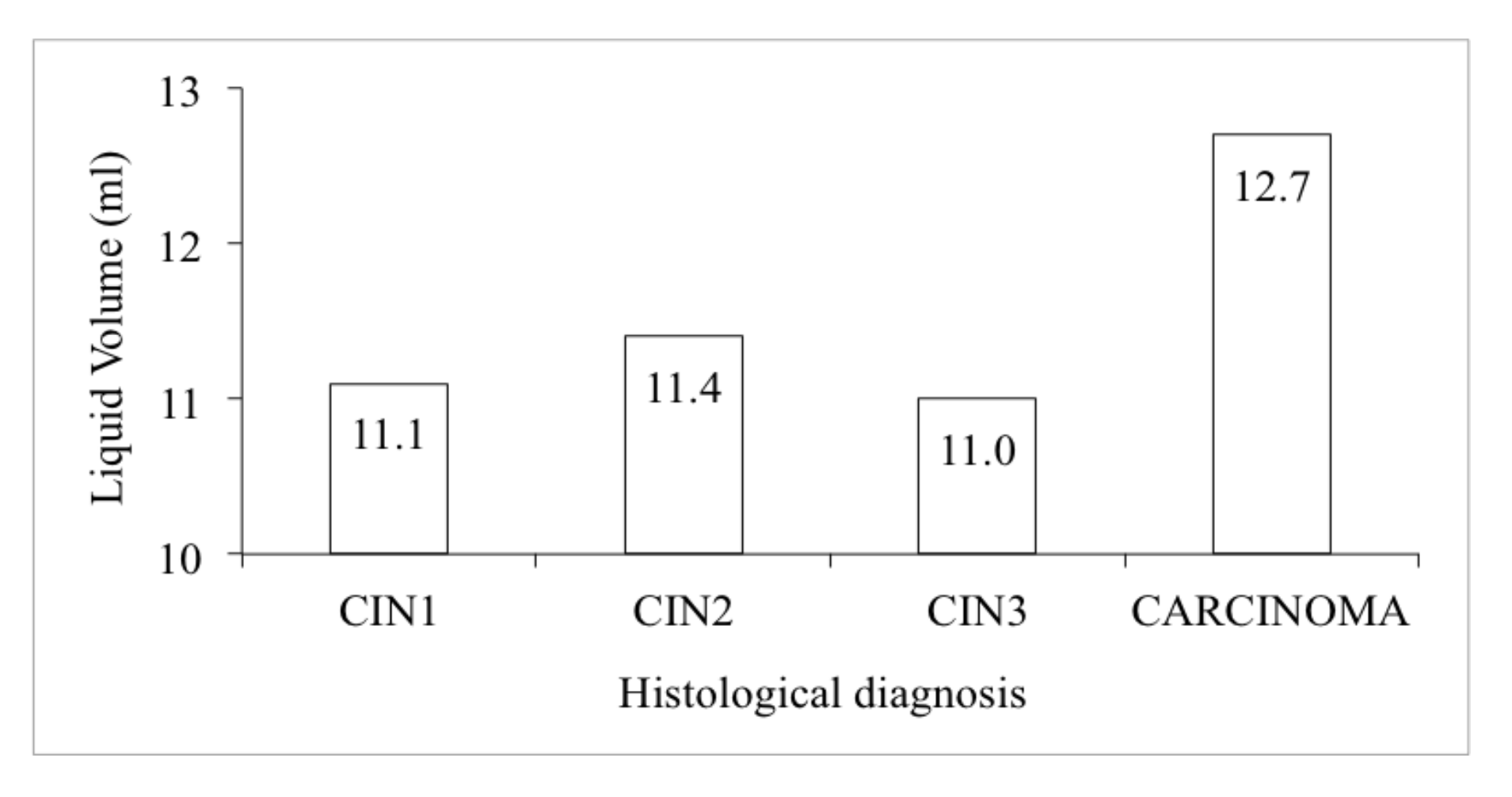
| HEPIKA Test | No. Cases | Histology | Residual Liquid Volume (mL) | Total Cells No. Estimation | Atypical Cells No. Estimation (%) | Optical Density E6#p53 | Optical Density E7#pRb |
|---|---|---|---|---|---|---|---|
| POSITIVE (27 Total) | 4 | ADC | 13 | 151,287 | 47,300 (43%) | 0.2985 | 0.2534 |
| 4 | SCC | 12 | 78,271 | 38,505 (58%) | 0.4731 | 0.3297 | |
| 9 | CIN3 | 13 | 142,661 | 24,224 (17%) | 0.2547 | 0.1993 | |
| 7 | CIN2 | 13 | 140,494 | 15,473 (12%) | 0.2190 | 0.2136 | |
| 3 | CIN1 | 8 | 138,842 | 14,055 (17%) | 0.1878 | 0.2133 | |
| Total | 12 | 133,413 | 26,360 (26%) | 0.2769 | 0.2319 | ||
| NEGATIVE (82 Total) | 1 | SCC | 15 | 47,235 | 33,508 (71%) | 0.1308 | 0.1430 |
| 1 | AIS | 15 | 219,621 | 173,463 (79%) | 0.2456 | 0.0967 | |
| 34 | CIN3 | 10 | 89,499 | 22,643 (26%) | 0.1015 | 0.1127 | |
| 25 | CIN2 | 11 | 98,123 | 16,122 (15%) | 0.1018 | 0.1099 | |
| 21 | CIN1 | 11 | 140,874 | 12,399 (9%) | 0.1027 | 0.1104 | |
| Total | 11 | 106,357 | 20,003 (19%) | 0.1040 | 0.1114 |
| Cytology | No. Cases | Hepika | HK+ | HK− | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | % | NEG | % | HPV+ | % | HPV− | % | HPV+ | % | HPV− | % | ||
| ADC | 0 | 2 | 0.7% | 2 | 100.0% | ||||||||
| SCC | 2 | 2 | 0.7% | 3 | 1.1% | 2 | 40.0% | 3 | 60.0% | ||||
| HSIL | 5 | 15 | 5.4% | 77 | 27.7% | 15 | 16.3% | 77 | 83.7% | ||||
| LSIL | 92 | 8 | 2.9% | 56 | 20.1% | 8 | 12.5% | 53 | 82.8% | 3 | 4.7% | ||
| ASC-H | 64 | 2 | 0.7% | 8 | 2.9% | 2 | 20.0% | 8 | 80.0% | ||||
| ASC-US | 10 | 2 | 0.7% | 2 | 100.0% | ||||||||
| AGC | 2 | 1 | 0.4% | 1 | 0.4% | 1 | 50.0% | 1 | 50.0% | ||||
| NEG | 35 | 7 | 2.5% | 28 | 10.1% | 7 | 20.0% | 28 | 80.0% | ||||
| N.P. | 66 | 3 | 1.1% | 63 | 22.7% | 3 | 4.5% | 63 | 95.5% | ||||
| Total | 210 | 37 | 14.4% | 175 | 85.6% | 37 | 13.3% | 3 | 1.1% | 172 | 61.9% | 3 | 23.7% |
| Histology | No. Cases | % | Hepika | HK+ | HK− | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | % | NEG | % | HPV+ | % | HPV− | % | HPV+ | % | HPV− | % | |||
| ADC | 0 | 1.5% | 4 | 100.0% | 4 | 100.0% | ||||||||
| SCC | 4 | 2.6% | 5 | 71.4% | 2 | 28.6% | 4 | 57.1% | 1 | 14.3% | 2 | 28.6% | ||
| CIN3–AIS | 61 | 22.3% | 11 | 18.0% | 50 | 82.0% | 11 | 18.0% | 50 | 82.0% | ||||
| CIN2 | 51 | 18.6% | 8 | 15.7% | 43 | 84.3% | 8 | 15.7% | 43 | 84.3% | ||||
| CIN1 | 34 | 12.4% | 7 | 26.6% | 27 | 79.4% | 7 | 20.6% | 27 | 79.4% | ||||
| NEG | 117 | 42.7% | 5 | 4.3% | 112 | 95.7% | 3 | 2.6% | 2 | 1.7% | 46 | 39.3% | 66 | 56.4% |
| Total | 274 | 100.0% | 40 | 14.6% | 234 | 85.4% | 37 | 13.5% | 3 | 1.1% | 168 | 61.3% | 66 | 24.1% |
| Histology | Sensitivity [% (95% CI)] | Specificity [% (95% CI)] | PPV [% (95% CI)] | NPV [% (95% CI)] | LR+ [% (95% CI)] | LR− [% (95% CI)] | Prevalence |
|---|---|---|---|---|---|---|---|
| CARCINOMA | 81.8% (48%–98%) | 88.2% (84%–92%) | 22.5% (16%–31%) | 99.1% (97%–100%) | 6.94 (4.50–10.70) | 0.21 (0.06–0.72) | 4.0% |
| CIN3+ | 27.8% (18%–40%) | 90.1% (85%–94%) | 50.0% (36%–64%) | 77.8% (75%–80%) | 2.81 (1.61–4.90) | 0.80 (0.69–0.93) | 26.3% |
| CIN2+ | 22.8% (16%–31%) | 92.1% (86%–96%) | 70.0% (55%–81%) | 59.4% (57%–62%) | 2.86 (1.52–5.39) | 0.84 (0.75–0.93) | 44.9% |
| Hepika POS | DOR | p Value | ||
|---|---|---|---|---|
| Histology | No. Cases | % | (95% CI) | |
| CARCINOMA | 9 | 81.8 (9/11) | 33.68 (6.95–163.07) | p < 0.001 |
| CIN3+ | 20 | 27.8 (20/72) | 3.50 (1.75–6.99) | p < 0.001 |
| CIN2+ | 28 | 22.8 (28/123) | 3.41 (1.65–7.05) | p < 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gustinucci, D.; Ciccocioppo, L.; Coppola, L.; Negri, G.; Zannoni, G.; Passamonti, B.; Cesarini, E.; Ianzano, C.; Andreano, T.; Pireddu, A.; et al. Multicentre Evaluation of Hepika Test Clinical Accuracy in Diagnosing HPV-Induced Cancer and Precancerous Lesions of the Uterine Cervix. Diagnostics 2021, 11, 619. https://doi.org/10.3390/diagnostics11040619
Gustinucci D, Ciccocioppo L, Coppola L, Negri G, Zannoni G, Passamonti B, Cesarini E, Ianzano C, Andreano T, Pireddu A, et al. Multicentre Evaluation of Hepika Test Clinical Accuracy in Diagnosing HPV-Induced Cancer and Precancerous Lesions of the Uterine Cervix. Diagnostics. 2021; 11(4):619. https://doi.org/10.3390/diagnostics11040619
Chicago/Turabian StyleGustinucci, Daniela, Lucia Ciccocioppo, Luigi Coppola, Giovanni Negri, Gianfranco Zannoni, Basilio Passamonti, Elena Cesarini, Ciro Ianzano, Tiziana Andreano, Anjuta Pireddu, and et al. 2021. "Multicentre Evaluation of Hepika Test Clinical Accuracy in Diagnosing HPV-Induced Cancer and Precancerous Lesions of the Uterine Cervix" Diagnostics 11, no. 4: 619. https://doi.org/10.3390/diagnostics11040619
APA StyleGustinucci, D., Ciccocioppo, L., Coppola, L., Negri, G., Zannoni, G., Passamonti, B., Cesarini, E., Ianzano, C., Andreano, T., Pireddu, A., & Giorgi-Rossi, P. (2021). Multicentre Evaluation of Hepika Test Clinical Accuracy in Diagnosing HPV-Induced Cancer and Precancerous Lesions of the Uterine Cervix. Diagnostics, 11(4), 619. https://doi.org/10.3390/diagnostics11040619






