CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
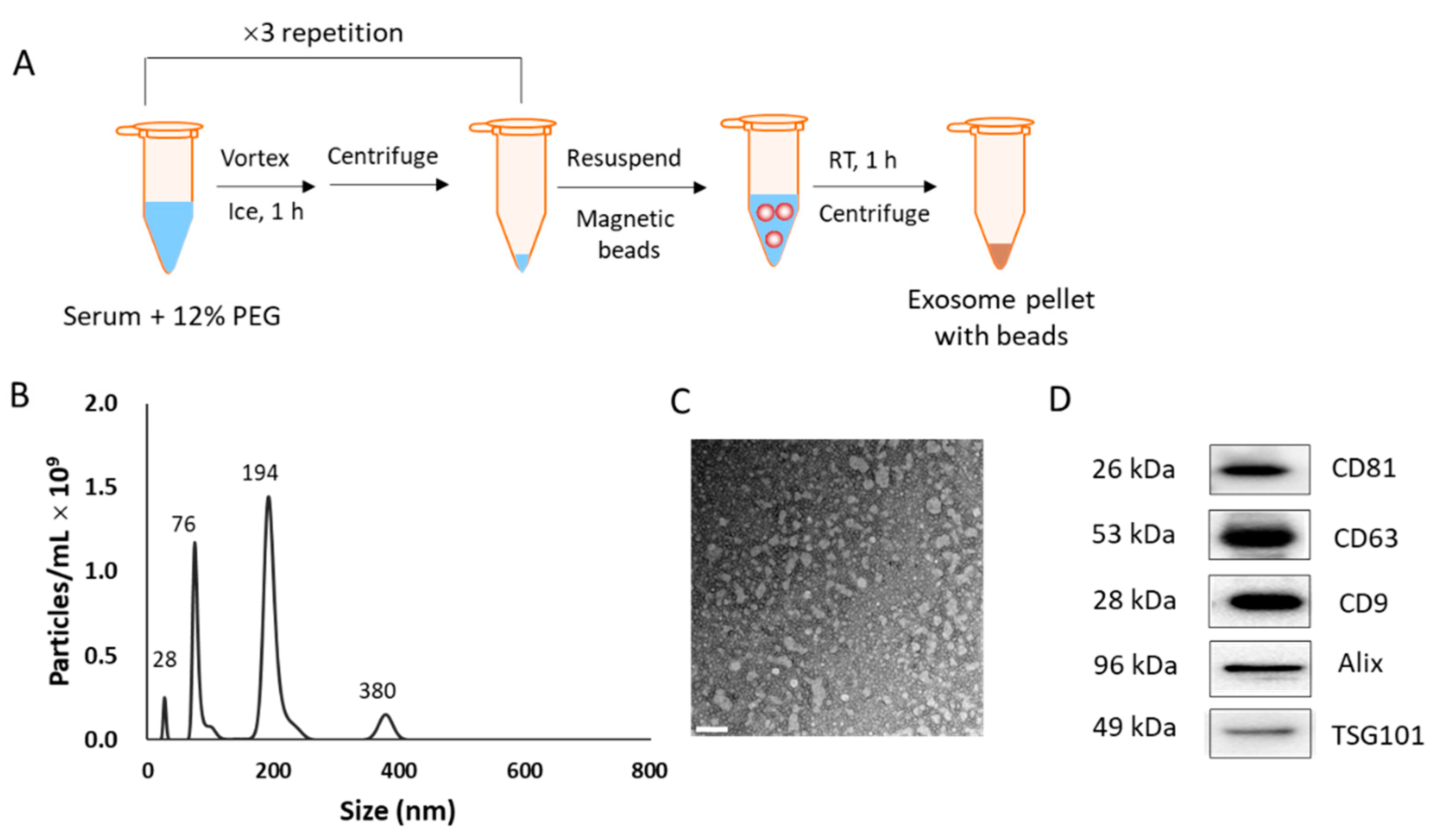
2.2. Isolation of EVs from Human Sera and Cell Culture Media
2.3. Characterization of EVs
2.4. Two-Dimensional Gel Electrophoresis of EV-Derived Proteins
2.5. Identification of Candidate Biomarkers
2.6. Confirmation of Biomarker Expression in EVs and Tissues
2.7. Pathway Analysis of Candidate Biomarkers
2.8. Transfection of Lung Cells with CD5L siRNA
2.9. Analysis of mRNA Expression
2.10. Statistical Analysis
3. Results
3.1. Isolation of EVs from Human Serum Samples and Subsequent Characterization
3.2. Protein Profiling for Lung Cancer-Derived EVs
3.3. Confirmation of Biomarker Candidates by Western Blotting
3.4. Expression of EV Proteins in Cancer Tissues
3.5. Biological Networks Related to the Biomarkers
3.6. Correlation of CD5L Level in EVs and Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, T.; Yang, X.; Huang, Y.; Zhao, M.; Li, M.; Ma, K.; Yin, J.; Zhan, C.; Wang, Q. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag. Res. 2019, 11, 943–953. [Google Scholar] [CrossRef] [Green Version]
- Knight, B.S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef] [Green Version]
- Del Ciello, A.; Franchi, P.; Contegiacomo, A.; Cicchetti, G.; Bonomo, L.; Larici, A.R. Missed lung cancer: When, where, and why? Diagn. Interv. Radiol. 2017, 23, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-based cancer biomarkers in liquid biopsy: A promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Hu, S.; Zhang, L.; Xin, J.; Sun, C.; Wang, L.; Ding, K.; Wang, B. Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis. Theranostics 2020, 10, 4544–4556. [Google Scholar] [CrossRef] [PubMed]
- Revelo, A.E.; Martin, A.; Velasquez, R.; Kulandaisamy, P.C.; Bustamante, J.; Keshishyan, S.; Otterson, G. Liquid biopsy for lung cancers: An update on recent developments. Ann. Transl. Med. 2019, 7, 349. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee, Advisory Boards, and Participating Institutions; International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee Advisory Boards and Participating Institutions. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, T.A.; Aae, T.F.; Brinchmann, J.E. Robust profiling of microRNAs and isomiRs in human plasma exosomes across 46 individuals. Sci. Rep. 2019, 9, 19999. [Google Scholar] [CrossRef]
- Yang, J.; Hagen, J.; Guntur, K.V.; Allette, K.; Schuyler, S.; Ranjan, J.; Petralia, F.; Gesta, S.; Sebra, R.; Mahajan, M.; et al. A next generation sequencing based approach to identify extracellular vesicle mediated mRNA transfers between cells. BMC Genom. 2017, 18, 987. [Google Scholar] [CrossRef]
- Jedinak, A.; Loughlin, K.R.; Moses, M.A. Approaches to the discovery of non-invasive urinary biomarkers of prostate cancer. Oncotarget 2018, 9, 32534–32550. [Google Scholar] [CrossRef]
- Oliveira, B.M.; Coorssen, J.R.; Martins-de-Souza, D. 2DE: The Phoenix of Proteomics. J. Proteom. 2014, 104, 140–150. [Google Scholar] [CrossRef]
- Doll, S.; Burlingame, A.L. Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 2015, 10, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Zhan, X.; Li, N.; Zhan, X.; Qian, S. Revival of 2DE-LC/MS in proteomics and its potential for large-scale study of human proteoforms. Med One 2018, 3, e180008. [Google Scholar]
- Sanjurjo, L.; Aran, G.; Roher, N.; Valledor, A.F.; Sarrias, M.-R. AIM/CD5L: A key protein in the control of immune homeostasis and inflammatory disease. J. Leukoc. Biol. 2015, 98, 173–184. [Google Scholar] [CrossRef] [Green Version]
- Oros, D.; Ceprnja, M.; Zucko, J.; Cindric, M.; Hozic, A.; Skrlin, J.; Barisic, K.; Melvan, E.; Uroic, K.; Kos, B.; et al. Identification of pathogens from native urine samples by MALDI–TOF/TOF tandem mass spectrometry. Clin. Proteom. 2020, 17, 25. [Google Scholar] [CrossRef]
- Chen, C.; Cui, S.; Li, W.; Jin, H.; Fan, J.; Sun, Y.; Cui, Z. Ingenuity pathway analysis of human facet joint tissues: Insight into facet joint osteoarthritis. Exp. Ther. Med. 2020, 19, 2997–3008. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Al Faruque, H.; Choi, E.-S.; Lee, H.-R.; Kim, J.-H.; Park, S.; Kim, E. Targeted removal of leukemia cells from the circulating system by whole-body magnetic hyperthermia in mice. Nanoscale 2020, 12, 2773–2786. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicle 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marmé, F.; Sültmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446. [Google Scholar] [CrossRef]
- Zheng, H.; Zhan, Y.; Liu, S.; Lu, J.; Luo, J.; Feng, J.; Fan, S. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J. Exp. Clin. Cancer Res. 2018, 37, 226. [Google Scholar] [CrossRef]
- Tsuno, H.; Arito, M.; Suematsu, N.; Sato, T.; Hashimoto, A.; Matsui, T.; Omoteyama, K.; Sato, M.; Okamoto, K.; Tohma, S.; et al. A proteomic analysis of serum-derived exosomes in rheumatoid arthritis. BMC Rheumatol. 2018, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Yan, Z.; Zeng, X.; Zhang, S.; Jiang, H.; Huang, H.; Zhuo, H. Serum and tissue proteomic signatures of patients with hepatocellular carcinoma using 2-D gel electrophoresis. Mol. Med. Rep. 2019, 20, 1025–1038. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.S.; Moraes, M.C.S.; Na, C.H.; Fierro-Monti, I.; Henriques, A.; Zahedi, S.; Bodo, C.; Tranfield, E.M.; Sousa, A.L.; Farinho, A.; et al. Is the proteome of bronchoalveolar lavage extracellular vesicles a marker of advanced lung cancer? Cancers 2020, 12, 3450. [Google Scholar] [CrossRef]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.-L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [Green Version]
- Li, S.P.; Lin, Z.X.; Jiang, X.Y.; Yu, X.-Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef] [Green Version]
- Ageta, H.; Ageta-Ishihara, N.; Hitach, K.; Karayel, O.; Onouchi, T.; Yamaguchi, H.; Kahyo, T.; Hatanaka, K.; Ikegami, K.; Yoshioka, Y.; et al. UBL3 modification influences protein sorting to small extracellular vesicles. Nat. Commun. 2018, 9, 3936. [Google Scholar] [CrossRef]
- Asada, H.; Tomiyasu, H.; Uchikai, T.; Ishihara, G.; Goto-Koshino, Y.; Ohno, K.; Tsujimoto, H. Comprehensive analysis of miRNA and protein profiles within exosomes derived from canine lymphoid tumour cell lines. PLoS ONE 2019, 14, e0208567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Huo, Z.; Nguyen, K.; Zhu, F.; Underwood, P.W.; Green Basso, K.B.; George, T.J.; Hughes, S.J. The proteome of pancreatic cancer-derived exosomes reveals signatures rich in key signaling pathways. Proteomics 2019, 19, 1800394. [Google Scholar] [CrossRef] [PubMed]
- Yuana, Y.; Levels, J.; Grootemaat, A.; Sturk, A.; Nieuwland, R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J. Extracell. Vesicles 2014, 8, 3. [Google Scholar] [CrossRef]
- Kim, M.H.; de Beer, M.C.; Wroblewski, J.M.; Charnigo, R.J.; Ji, A.; Webb, N.R.; de Beer, F.C.; van der Westhuyzen, D.R. Impact of individual acute phase serum amyloid A isoforms on HDL metabolism in mice. J. Lipid Res. 2016, 57, 969–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindy, M.S.; de Beer, M.C.; Yu, J.; de Beer, F.C. Expression of mouse acute-phase (SAA1.1) and constitutive (SAA4) serum amyloid A isotypes: Influence on lipoprotein profiles. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1543–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, N.; Hatano, E.; Iguchi, K.; Sato, M.; Kawaguchi, H.; Ohtsu, I.; Sakurai, T.; Aizawa, N.; Iijima, H.; Nishiguchi, S.; et al. Elevated levels of circulating ITIH4 are associated with hepatocellular carcinoma with nonalcoholic fatty liver disease: From pig model to human study. BMC Cancer 2019, 19, 621. [Google Scholar] [CrossRef]
- Vaisar, T.; Pennathur, S.; Green, P.S.; Gharib, S.A.; Hoofnagle, A.N.; Cheung, M.C.; Byun, J.; Vuletic, S.; Kassim, S.; Singh, P.; et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 2007, 117, 746–756. [Google Scholar] [CrossRef]
- Wessel, H.; Saeed, A.; Heegsma, J.; Connelly, M.A.; Faber, K.N.; Dullaart, R.P. Plasma levels of retinol binding protein 4 relate to large VLDL and small LDL particles in subjects with and without type 2 diabetes. J. Clin. Med. 2019, 8, 1792. [Google Scholar] [CrossRef] [Green Version]
- Speeckaert, M.M.; Taes, Y.E.; De Buyzere, M.L.; Christophe, A.B.; Kaufman, J.-M.; Delanghe, J.R. Investigation of the potential association of vitamin D binding protein with lipoproteins. Ann. Clin. Biochem. 2010, 47, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Han, H.; Yan, X.; Ding, F.; Su, X.; Wang, H.; Chen, Q.; Lu, L.; Zhang, R.; Jin, W. Tetranectin as a potential biomarker for stable coronary artery disease. Sci. Rep. 2015, 5, 17632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plubell, D.L.; Fenton, A.M.; Rosario, S.; Bergstrom, P.; Wilmarth, P.A.; Clark, W.M.; Zakai, N.A.; Quinn, J.F.; Minnier, J.; Alkayed, N.J.; et al. High-density lipoprotein carries markers that track with recovery from stroke. Circ. Res. 2020, 127, 1274–1287. [Google Scholar] [CrossRef]
- Nikitidou, E.; Khoonsari, P.E.; Shevchenko, G.; Ingelsson, M.; Kultima, K.; Erlandsson, A. Increased release of apolipoprotein E in extracellular vesicles following amyloid-β protofibril exposure of neuroglial co-cultures. J. Alzheimers Dis. 2017, 60, 305–321. [Google Scholar] [CrossRef] [Green Version]
- Tutanov, O.; Orlova, E.; Proskura, K.; Grigor’eva, A.; Yunusova, N.; Tsentalovich, Y.; Alexandrova, A.; Tamkovich, S. Proteomic analysis of blood exosomes from healthy females and breast cancer patients reveals an association between different exosomal bioactivity on non-tumorigenic epithelial cell and breast cancer cell migration in vitro. Biomolecules 2020, 10, 495. [Google Scholar] [CrossRef] [Green Version]
- Kurokawa, J.; Arai, S.; Nakashima, K.; Nagano, H.; Nishijima, A.; Miyata, K.; Ose, R.; Mori, M.; Kubota, N.; Kadowaki, T.; et al. Macrophage-derived AIM is endocytosed into adipocytes and decreases lipid droplets via inhibition of fatty acid synthase activity. Cell Metab. 2010, 11, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Qu, P.; Wu, L.; Li, B.; Du, H.; Yan, C. Api6/AIM/Spα/CD5L overexpression in alveolar type II epithelial cells induces spontaneous lung adenocarcinoma. Cancer Res. 2011, 71, 5488–5499. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Yan, X.; Yin, Y.; Lin, X.; Zhang, Q.; Xia, Y.; Cao, J. Therapeutic targeting of apoptosis inhibitor of macrophage/CD5L in sepsis. Am. J. Respir. Cell Mol. Biol. 2019, 60, 323–334. [Google Scholar] [CrossRef]





| Disease Type | No. of Samples | Age (Mean, Range) | Sex (Male/Female) | Number of Samples (Stage I, II, III, IV) |
|---|---|---|---|---|
| Normal | 20 | 50 (47–63) | 7/13 | - |
| Lung cancer | ||||
| AC | 20 | 65 (53–81) | 10/10 | 6, 0, 5, 9 |
| SCC | 20 | 72 (52–83) | 16/4 | 1, 0, 11, 8 |
| SCLC | 20 | 70 (52–82) | 18/2 | 0, 1, 6, 13 |
| Total | 60 | 69 (52–83) | 44/16 | 7, 1, 22, 30 |
| Protein | Symbol | AUC | Sensitivity | Specificity | SCLC * | AC * | SCC * | PanC * | CRC * |
|---|---|---|---|---|---|---|---|---|---|
| CD5 antigen-like | CD5L | 0.943 | 92.9 | 94.1 | 4.4 | 4.1 | 4.0 | 0.3 | 0.4 |
| Retinol binding protein 4 | RBP4 | 0.917 | 90.5 | 88.2 | 13.0 | 22.8 | 18.7 | 0.1 | 0.1 |
| Serum amyloid A beta | SAA1 | 0.893 | 78.6 | 100.0 | 18.3 | 115.0 | 168.5 | 1.0 | 1.0 |
| Tetranectin | CLEC3B | 0.887 | 88.1 | 76.5 | 3.3 | 16.7 | 9.0 | 0.0 | 0.0 |
| Inter-alpha (globulin) inhibitor H4 | ITIH4 | 0.873 | 81.0 | 88.2 | 9.2 | 7.5 | 4.8 | 0.0 | 0.0 |
| Serpin peptidase inhibitor, clade F | SERPINF1 | 0.833 | 83.3 | 76.5 | 2.1 | 2.5 | 2.3 | 0.5 | 0.5 |
| Serum amyloid A-4 | SAA4 | 0.833 | 66.7 | 100.0 | 22.2 | 8.7 | 29.2 | 1.0 | 1.0 |
| Serpin peptidase inhibitor, clade C | SERPINC1 | 0.824 | 71.4 | 88.2 | 10.2 | 10.8 | 15.4 | 0.5 | 0.5 |
| Vitamin D binding protein | DBP | 0.798 | 59.5 | 100.0 | 131.5 | 136.0 | 79.9 | 1.0 | 1.0 |
| Chromosome 20 open reading frame 3 | C20ORF3 (APMAP) | 0.753 | 73.8 | 82.4 | 2.9 | 2.8 | 2.5 | 0.0 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, E.-S.; Faruque, H.A.; Kim, J.-H.; Kim, K.J.; Choi, J.E.; Kim, B.A.; Kim, B.; Kim, Y.J.; Woo, M.H.; Park, J.Y.; et al. CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer. Diagnostics 2021, 11, 620. https://doi.org/10.3390/diagnostics11040620
Choi E-S, Faruque HA, Kim J-H, Kim KJ, Choi JE, Kim BA, Kim B, Kim YJ, Woo MH, Park JY, et al. CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer. Diagnostics. 2021; 11(4):620. https://doi.org/10.3390/diagnostics11040620
Chicago/Turabian StyleChoi, Eun-Sook, Hasan Al Faruque, Jung-Hee Kim, Kook Jin Kim, Jin Eun Choi, Bo A. Kim, Bora Kim, Ye Jin Kim, Min Hee Woo, Jae Yong Park, and et al. 2021. "CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer" Diagnostics 11, no. 4: 620. https://doi.org/10.3390/diagnostics11040620
APA StyleChoi, E.-S., Faruque, H. A., Kim, J.-H., Kim, K. J., Choi, J. E., Kim, B. A., Kim, B., Kim, Y. J., Woo, M. H., Park, J. Y., Hur, K., Lee, M.-Y., Kim, D. S., Lee, S. Y., & Kim, E. (2021). CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer. Diagnostics, 11(4), 620. https://doi.org/10.3390/diagnostics11040620







