The Prognostic and Predictive Role of Somatic BRCA Mutations in Ovarian Cancer: Results from a Multicenter Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. BRCA Somatic Testing and Variant Interpretation
2.3. Statistical Analysis
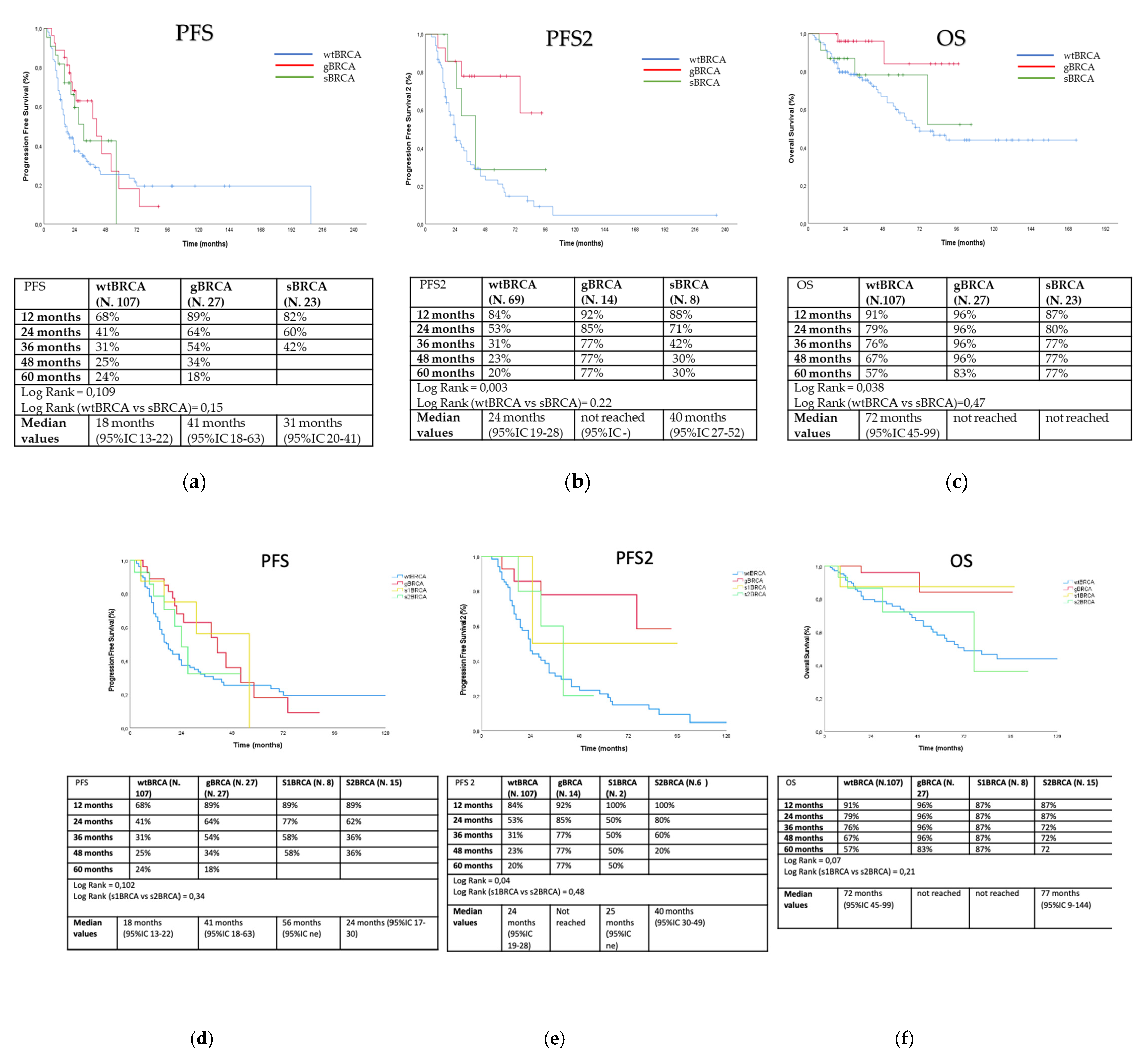
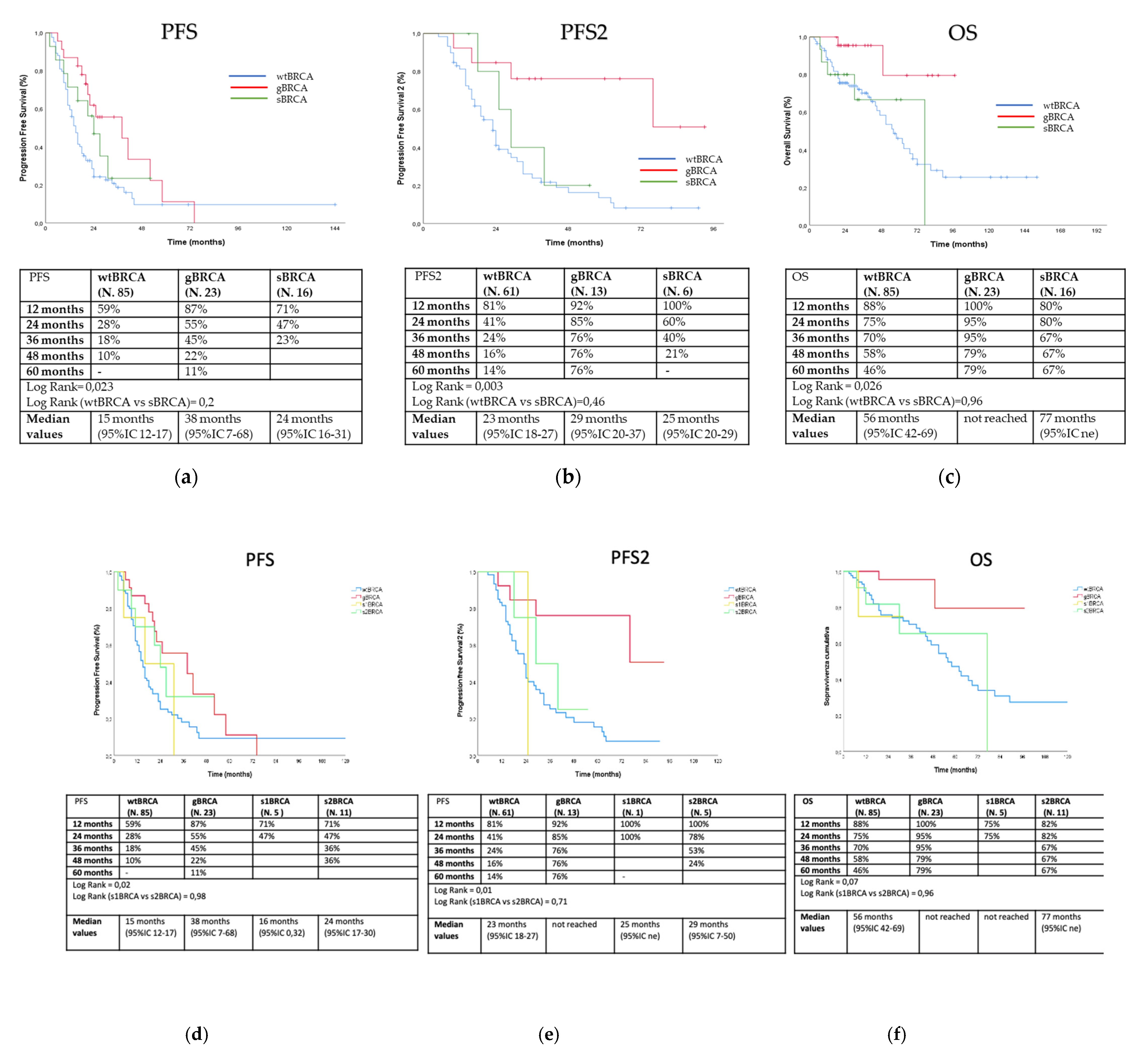
3. Results
3.1. Patient Characteristics
3.2. Outcome Analysis
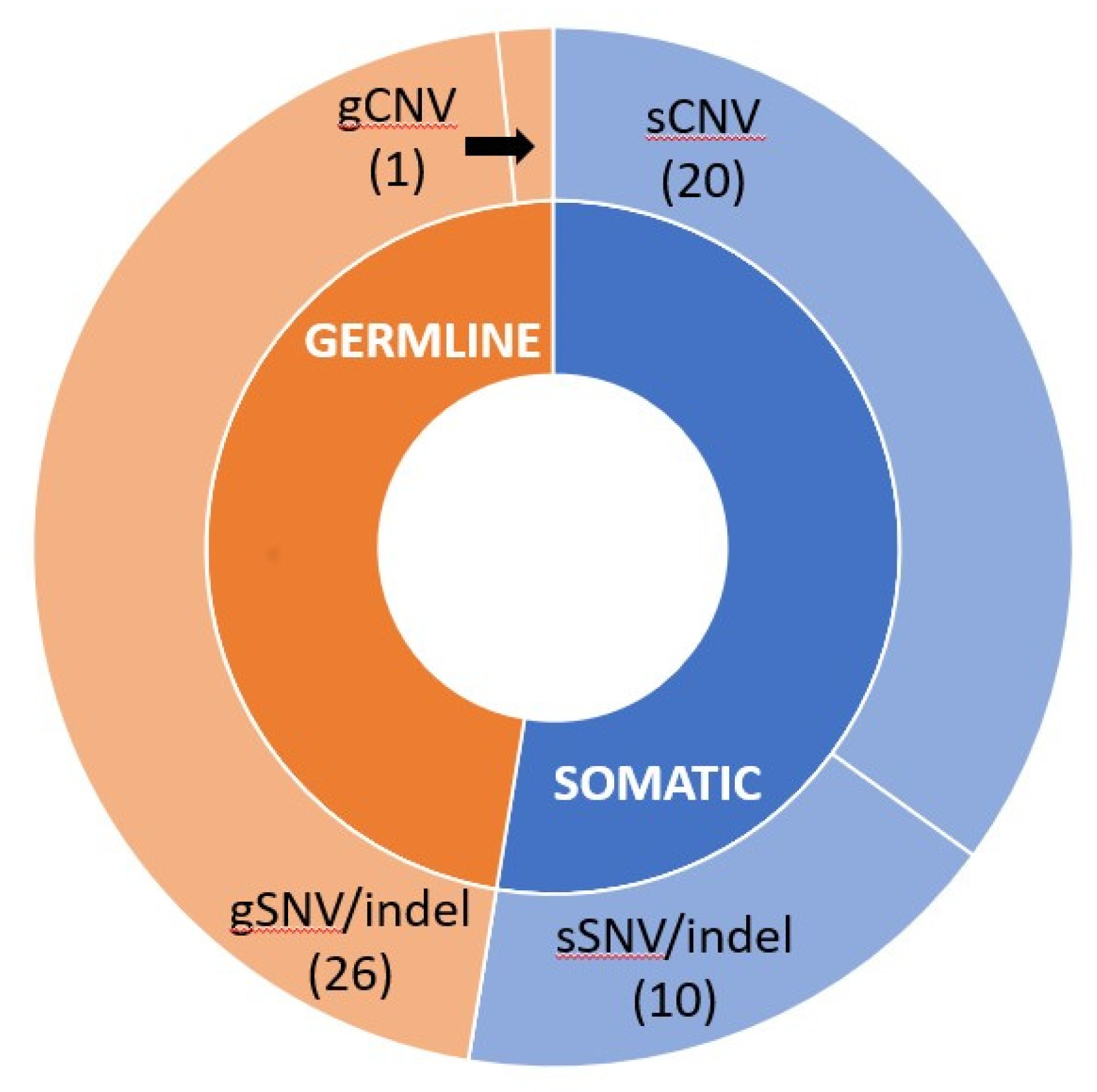
3.3. Detected BRCA Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Toss, A.; Molinaro, E.; Sammarini, M.; Del Savio, M.C.; Cortesi, L.; Facchinetti, F.; Grandi, G. Hereditary ovarian cancers: State of the art. Minerva Med. 2019, 110, 301–319. [Google Scholar] [CrossRef]
- Alsop, K.; Fereday, S.; Meldrum, C.; DeFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012, 30, 2654–2663. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D. Somatic mutations in BRCA1 andBRCA2 could expand the number of patients that benefit from poly (ADPribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 2010, 28, 3570–3576. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, C.E.; van Ierland, Y.; van Asperen, C.J.; Hallensleben, E.; Devilee, P.; Feluren, G.J.; Kenter, G.G. Prediction of BRCA1/2 mutation status in patients with ovarian cancer from a hospital-based cohort. Genet Med. 2007, 9, 173–179. [Google Scholar] [CrossRef]
- Malander, S.; Ridderheim, M.; Masback, A.; Loman, N.; Kristoffersson, U.; Olsson, H.; Nibert, M.; Borg, A. One in 10 ovarian cancer patients carry germ line BRCA1 or BRCA2 mutations: Results of a prospective study in Southern Sweden. Eur. J. Cancer 2004, 40, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Risch, N. The genetic epidemiology of cancer: Interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol. Biomarkers Prev. 2001, 10, 733–741. [Google Scholar]
- Soegaard, M.; Kjaer, S.K.; Cox, M.; Wozniak, E.; Hogdall, E.; Hogdall, C.; Blaakaer, J.; Jacobs, I.J.; Gayther, S.A.; Ramus, S.J. BRCA1 and BRCA2 mutation prevalence and clinical characteristics of a population-based series of ovarian cancer cases from Denmark. Clin. Cancer Res. 2008, 14, 3761–3767. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, A.V.; Fostira, F.; Pertesi, M.; Tsitlaidou, M.; Voutsinas, G.E.; Triantafyllidou, O.; Bamias, A.; Dimopoulos, M.A.; Timotheadou, E.; Pectasides, D.; et al. Prevalence of BRCA1 mutations in familial and sporadic Greek ovarian cancer cases. PLoS ONE 2013, 8, e58182. [Google Scholar] [CrossRef]
- Ben David, Y.; Chetrit, A.; Hirsh-Yechezkel, G.; Friedman, E.; Beck, B.D.; Beller, U.; Ben-Baruch, G.; Fishman, A.; Levavi, H.; Lubin, F.; et al. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J. Clin. Oncol. 2002, 20, 463–466. [Google Scholar] [CrossRef]
- Bolton, K.L.; Chenevix-Trench, G.; Goh, C.; Sadetzki, S.; Ramus, S.J.; Karlan, B.Y.; Lambrechts, D.; Despierre, E.; Barrowdale, D.; McGuffog, L.; et al. Association between BRCA1 andBRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012, 307, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.; Sonoda, Y.; Federici, M.G.; Bogomolniy, F.; Rhei, E.; Maresco, D.L.; Saigo, P.E.; Almadrones, L.A.; Barakat, P.R.; Brown, C.L.; et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA 2000, 283, 2260–2265. [Google Scholar] [CrossRef] [PubMed]
- Cass, I.; Baldwin, R.L.; Varkey, T.; Moslehi, R.; Narod, S.A.; Karlan, B.Y. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 2003, 97, 2187–2195. [Google Scholar] [CrossRef]
- Chetrit, A.; Hirsh-Yechezkel, G.; Ben-David, Y.; Lubin, F.; Friedman, E.; Sadetzki, S. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: The national Israeli study of ovarian cancer. J. Clin. Oncol. 2008, 26, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Majdak, E.J.; Debniak, J.; Milczek, T.; Cornelisse, C.J.; Devilee, P.; Emerich, J.; Jassem, J.; De Bock, G.H. Prognostic impact of BRCA1 pathogenic and BRCA1/BRCA2 unclassified variant mutations in patients with ovarian carcinoma. Cancer 2005, 104, 1004–1012. [Google Scholar] [CrossRef]
- Yang, D.; Khan, S.; Sun, Y.; Hess, K.; Shmulevich, I.; Sood, A.K.; Zhang, W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011, 306, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Toss, A.; Cortesi, L. Molecular Mechanisms of PARP Inhibitors in BRCA-related Ovarian Cancer. J. Cancer Sci. Ther. 2013, 5, 409–416. [Google Scholar]
- Kurian, A.W.; Kingham, K.E.; Ford, J.M. Next-generation sequencing for hereditary breast and gynecologic cancer risk assessment. Curr. Opin. Obstet. Gynecol. 2015, 27, 23–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Axilbund, J.E. Panel testing is not a panacea. J. Clin. Oncol. 2016, 34, 1433–1435. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.A.; Anderson, M.E.; Lacour, R.A.; Suri, A.; Daniels, M.S.; Urbauer, D.L.; Nogueras-Gonzalez, G.M.; Scmeler, K.M.; Gershenson, D.M.; Lu, K.H. Evaluating women with ovarian cancer for BRCA1 and BRCA2 mutations: Missed opportunities. Obstet. Gynecol. 2010, 115, 945–952. [Google Scholar] [CrossRef]
- Randall, L.M.; Pothuri, B.; Swisher, E.M.; Diaz, J.P.; Buchanan, A.; Witkop, C.T.; Powell, C.B.; Smith, E.B.; Robson, M.E.; Boyd, J.; et al. Multi-disciplinary summit on genetics services for women with gynecologic cancers: A society of gynecologic oncology white paper. Gynecol. Oncol. 2017, 146, 217–224. [Google Scholar] [CrossRef]
- Hahnen, E.; Baumann, K.H.; Heimbach, A.; Reuss, A.; Jackisch, C.; Hauke, J.; Park-Simon, T.W.; Richters, L.K.; Hanker, L.C.; Kroeber, S.; et al. Prevalence of somatic mutations in risk genes including BRCA 1/2 in consecutive ovarian cancer patients (AGO-TR-1 study). J. Clin. Oncol. 2016, 34, 5544. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.B.; Banerjee, S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef]
- Lesnock, J.; Krivak, T.; Kiet, T.; Sherman, A.; Monk, B.; Ruskin, R.; Kapp, D.; Chen, L.; Chan, J. Germline versus Somatic BRCA mutations: Analysis of the cancer genome atlas project in patients with advanced stage serous epithelial ovarian cancer. Gynecol. Oncol. 2012, 125, S34. [Google Scholar] [CrossRef]
- George, A.; Banerjee, S.; Kaye, S. Olaparib and somatic BRCA mutations. Oncotarget 2017, 8, 43598–43599. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- You, Y.; Li, L.; Lu, J.; Wu, H.; Wang, J.; Gao, J.; Wu, M.; Liang, Z. Germline and Somatic BRCA1/2 Mutations in 172 Chinese Women with Epithelial Ovarian Cancer. Front. Oncol. 2020, 10, 295. [Google Scholar] [CrossRef] [PubMed]
- Italian Association of Medical Oncology. Raccomandazioni per l’implementazione del test BRCA nelle pazienti con carcinoma ovarico e nei familiari a rischio elevato di neoplasia. V.2-gennaio 2019. Available online: https://www.aiom.it/raccomandazioni-per-limplementazione-del-test-brca-nelle-pazienti-con-carcinoma-ovarico-e-nei-familiari-a-rischio-elevato-di-neoplasia/ (accessed on 5 October 2020).
- Gori, S.; Barberis, M.; Bella, M.A.; Buttitta, F.; Capoluongo, E.; Carrera, P.; Colombo, N.; Cortesi, L.; Genuardi, M.; Gion, M.; et al. Recommendations for the implementation of BRCA testing in ovarian cancer patients and their relatives. Crit. Rev. Oncol. Hematol. 2019, 140, 67–72. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib Maintenance Therapy in Platinum-Sensitive Relapsed Ovarian Cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef]
- Kaufman, B.; Shapira-Frommer, R.; Schmutzler, R.K.; Audeh, M.W.; Friedlander, M.; Balmaña, J.; Mitchell, G.; Fried, G.; Stemmer, S.M.; Hubert, A.; et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015, 33, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nuclei Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- McLaren, W.; Pritchard, B.; Rios, D.; Chen, Y.; Flicek, P.; Cunningham, F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010, 26, 2069–2070. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef]
- Choi, M.C.; Hwang, S.; Kim, S.; Jung, S.G.; Park, H.; Joo, W.D.; Song, S.H.; Lee, C.; Kim, T.H.; Kang, H.; et al. Clinical Impact of Somatic Variants in Homologous Recombination Repair-Related Genes in Ovarian High-Grade Serous Carcinoma. Cancer Res. Treat 2020, 52, 634–644. [Google Scholar] [CrossRef] [PubMed]
| BC and OC in the Same Patient or Family |
|---|
| OC, fallopian tube, or primary peritoneal cancer (mucinous and borderline types excluded) at any age |
| Male BC |
| Triple-negative BC diagnosed at ≤60 years |
| BC diagnosed at ≤35 years |
| At least 2 first-degree blood relatives with BC with at least 1 diagnosed ≤40 years or with a bilateral presentation in the same family |
| wtBRCA (n = 107, 67.7%) | gBRCA (n = 27, 17.1%) | sBRCA (n =24, 15.2%) | p Value | ||||
|---|---|---|---|---|---|---|---|
| Median age (SD) | 70.50 (0.71) | 58.50 (19.09) | 56.00 (16.97) | 0.04 | |||
| n | % | n | % | n | % | ||
| Family history of BC 1 | 38 | 35.51 | 14 | 51.85 | 9 | 37.50 | >0.05 |
| Family history of OC 1 | 7 | 6.54 | 6 | 22.22 | 4 | 16.67 | >0.05 |
| Family history of BC+OC 1 | 2 | 1.87 | 4 | 14.81 | 1 | 4.17 | >0.05 |
| FIGO stage | |||||||
| I | 11 | 10.28 | 2 | 7.41 | 6 | 25.00 | >0.05 |
| II | 12 | 11.21 | 1 | 3.70 | 2 | 8.33 | |
| Total I–II | 23 | 21.50 | 3 | 11.11 | 8 | 33.33 | |
| III | 50 | 46.73 | 18 | 66.67 | 13 | 54.17 | |
| IV | 33 | 30.84 | 5 | 18.52 | 3 | 12.50 | |
| Total III–IV | 83 | 77.57 | 23 | 85.19 | 16 | 66.67 | |
| Unknown | 1 | 0.93 | 1 | 3.70 | 0 | 0 | |
| Histotype | |||||||
| Serous | 85 | 79.44 | 25 | 92.59 | 19 | 79.17 | >0.05 |
| Mucinous | 2 | 1.87 | 0 | 0 | 0 | 0 | |
| Clear Cells | 4 | 3.74 | 0 | 0 | 3 | 12.50 | |
| Endometrioid | 16 | 14.95 | 1 | 3.70 | 2 | 8.33 | |
| Transitional | 0 | 0 | 1 | 3.70 | 0 | 0 | |
| Non-serous | 22 | 20.56 | 2 | 7.41 | 5 | 20.83 | |
| Surgery | |||||||
| No | 14 | 13.08 | 1 | 3.70 | 1 | 4.17 | >0.05 |
| Upfront | 66 | 61.68 | 16 | 59.26 | 19 | 79.17 | |
| Interval | 10 | 9.35 | 5 | 18.52 | 4 | 16.67 | |
| Post-neoadjuvant therapy | 17 | 15.89 | 5 | 18.52 | 0 | 0 | 0.05 |
| Residual | |||||||
| yes | 23 | 24.73 | 5 | 19.23 | 6 | 26.09 | >0.05 |
| no | 70 | 75.26 | 21 | 80.77 | 16 | 69.57 | |
| Unknown | 0 | 0 | 0 | 0 | 1 | 4.35 | |
| First line therapy | |||||||
| Platinum-based | 107 | 100 | 27 | 100 | 23 | 95.83 | >0.05 |
| Non-platinum-based | 0 | 0 | 0 | 0 | |||
| None | 0 | 0 | 1 | 4.17 |
| OS | PFS | ||||
|---|---|---|---|---|---|
| wtBRCA | wtBRCA | ||||
| Factor | Status | HR (95 CI) | p-Value | HR (95 CI) | p-Value |
| Age | <50 | 1.00 | 1.00 | ||
| ≥50 | 1.02 (0.78–1.56) | 0.9 | 0.24 (0.09–1.54) | 0.83 | |
| FIGO | 1–2 | 1.00 | 1.00 | ||
| 3–4 | 1.69 (1.18–10.8) | 0.04 | 0.17 (0.08–0.61) | 0.05 | |
| Serous histotype | No | 1.00 | 1.00 | ||
| Yes | 0.35 (0.18–1.01) | 0.26 | 1.32 (0.21–4) | 0.42 | |
| Surgery | Yes | 1.00 | 1.00 | ||
| No | 0.43 (0.07–0.99) | 0.001 | 1.34 (1.07–1.92) | 0.003 | |
| Residual | No | 1.00 | 1.00 | ||
| Yes | 0.31 (0.13–0.77) | 0.004 | 1.7 (1.02–2.4) | 0.01 | |
| Patient | BRCA1 | BRCA2 | VAF in the Tested Somatic Sample (%) | Copy Number (CN) Call in the Tested Somatic Sample | Confirmed in the Matched Germline Sample | sBRCA/ gBRCA |
|---|---|---|---|---|---|---|
| 1 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 2 | / | c.993_994delAAinsG,p.(Ile332PhefsX17) | 52.6 | no | somatic | |
| 3 | c.4675+1G>A | / | 5.4 | no | somatic | |
| 4 | / | / | / | BRCA1DEL Exons16-20; CN = 1 | no | somatic |
| 5 | / | c.8629G>T, p.(Glu2877Ter) | 16 | no | somatic | |
| 6 | / | c.5073dupA, p.(Trp1692Metfs*3) | 8.6 | no | somatic | |
| 7 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| 8 | c.331G>T, p.(Glu111Ter) | / | 50.2 | no | somatic | |
| 9 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| c.2269delG, p.Val757PhefsX8 | / | 59.2 | no | somatic | ||
| 10 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| 11 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 12 | c.1687C>T, p.(Gln563*) | / | 79.5 | no | somatic | |
| / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic | |
| 13 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| 14 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 15 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 16 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 17 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 18 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 19 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 20 | / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic |
| 21 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| 22 | c.2670delG, p.Ser891Profs*2 | / | 50.5 | no | somatic | |
| 23 | / | c.6611delC, p.(Pro2204Leufs*2) | 54.3 | no | somatic | |
| / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic | |
| 24 | / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic |
| 25 | c.3916_3917delTT, p.Leu1306AspfsX23 | / | 78.9 | yes | germline | |
| 26 | / | c.7975A>G, p.(Arg2659Gly) | 80 | yes | germline | |
| 27 | / | c.3847_3848delGT, p.(Val1283LysfsX2) | 85 | yes | germline | |
| 28 | / | c.6037A>T, p.(Lys2013Ter) | 85 | yes | germline | |
| 29 | c.547+2T>A | / | 83.4 | yes | germline | |
| 30 | c.2157_2160delAGAA, p.(Lys719AsnfsX16) | / | 92.8 | yes | germline | |
| 31 | c.4357+1delG | / | 90 | yes | germline | |
| 32 | c.3607C>T, p.(Arg1203Ter) | / | 59,2 | yes | germline | |
| 33 | c.4096+1G>A | / | 73.4 | yes | germline | |
| 34 | c.3481_3491delGAAGATACTAG, p.(Glu1161PhefsTer3) | / | 91.4 | yes | germline | |
| 35 | c.843_846delCTCA, p.(Ser282TyrX15) | / | 83.2 | yes | germline | |
| / | / | / | BRCA1DEL entire gene; CN = 1 | no | somatic | |
| 36 | c.3288_3289delAA, p.(Leu1098SerfsX4) | / | 94.6 | yes | germline | |
| 37 | c.5434C>G, p.(Pro1812Ala) | / | 63.8 | yes | germline | |
| 38 | / | c.9097dupA, p.(Thr3033AsnfsX11) | Tissue not available | yes | germline | |
| 39 | c.5017_5019delCAC, p.(His1673del) | / | 64.8 | yes | germline | |
| 40 | / | c.5722_5723delCT, p.(Leu1908Argfs*2) | 60.3 | yes | germline | |
| 41 | / | c.7180A>T, p.(Arg2394*) | 78.7 | yes | germline | |
| 42 | c.3916_3917delTT, p.(Leu1306Aspfs*23) | / | 76 | yes | germline | |
| 43 | c.4508C>A, p.(Ser1503*) | / | 88.6 | yes | germline | |
| 44 | c.4484G>T, p.(Arg1495Met) | / | 73 | yes | germline | |
| 45 | c.2157_2160delAGAA, p.(Lys719Asnfs*16) | / | 84.3 | yes | germline | |
| 46 | c.3979C>T, p.(Gln1327*) | / | 84.6 | yes | germline | |
| 47 | / | c.9154C>T, p.(Arg3052Trp) | 59.3 | yes | germline | |
| 48 | / | / | / | BRCA1DEL Promotor-Exon2; CN = 1 | yes | germline |
| 49 | / | c.3847_3848delGT, p.(Val1283Lysfs*2) | 77.4 | yes | germline | |
| / | / | / | BRCA2DEL entire gene; CN = 1 | no | somatic | |
| 50 | c.3916_3917delTT, p.(Leu1306Aspfs*23) | / | 70.2 | yes | germline | |
| 51 | / | c.7558C>T, p.(Arg2520*) | 5.8 | yes | germline |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toss, A.; Piombino, C.; Tenedini, E.; Bologna, A.; Gasparini, E.; Tarantino, V.; Filieri, M.E.; Cottafavi, L.; Giovanardi, F.; Madrigali, S.; et al. The Prognostic and Predictive Role of Somatic BRCA Mutations in Ovarian Cancer: Results from a Multicenter Cohort Study. Diagnostics 2021, 11, 565. https://doi.org/10.3390/diagnostics11030565
Toss A, Piombino C, Tenedini E, Bologna A, Gasparini E, Tarantino V, Filieri ME, Cottafavi L, Giovanardi F, Madrigali S, et al. The Prognostic and Predictive Role of Somatic BRCA Mutations in Ovarian Cancer: Results from a Multicenter Cohort Study. Diagnostics. 2021; 11(3):565. https://doi.org/10.3390/diagnostics11030565
Chicago/Turabian StyleToss, Angela, Claudia Piombino, Elena Tenedini, Alessandra Bologna, Elisa Gasparini, Vittoria Tarantino, Maria Elisabetta Filieri, Luca Cottafavi, Filippo Giovanardi, Stefano Madrigali, and et al. 2021. "The Prognostic and Predictive Role of Somatic BRCA Mutations in Ovarian Cancer: Results from a Multicenter Cohort Study" Diagnostics 11, no. 3: 565. https://doi.org/10.3390/diagnostics11030565
APA StyleToss, A., Piombino, C., Tenedini, E., Bologna, A., Gasparini, E., Tarantino, V., Filieri, M. E., Cottafavi, L., Giovanardi, F., Madrigali, S., Civallero, M., Marcheselli, L., Marchi, I., Domati, F., Venturelli, M., Barbieri, E., Grandi, G., Tagliafico, E., & Cortesi, L. (2021). The Prognostic and Predictive Role of Somatic BRCA Mutations in Ovarian Cancer: Results from a Multicenter Cohort Study. Diagnostics, 11(3), 565. https://doi.org/10.3390/diagnostics11030565







