Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis
Abstract
1. Introduction
2. Results
2.1. Characteristics of Study Subjects
2.2. Circulating miRNA Expression in Surgery Naïve Stage IV CRC Patients Compared to Healthy Subjects
2.3. Circulating miRNA Expression in Surgery Naïve vs. Postoperative and Recurrent Stage IV CRC Patients
2.4. Diagnostic Accuracy of miRNA for CRC Detection
2.5. Diagnostic Accuracy of miRNA for Liver Metastasis Detection
3. Discussion
4. Materials and Methods
4.1. Specimen Collection
4.2. Total RNA Extraction
4.3. miRNA Expression Using Reverse Transcription Quantitative Real Time Polymerase Chain Reaction (RT-qPCR)
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Khachfe, H.H.; Salhab, H.A.; Fares, M.Y.; Khachfe, H.M. Probing the Colorectal Cancer Incidence in Lebanon: An 11-Year Epidemiological Study. J. Gastrointest. Cancer 2020, 51, 805–812. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.K.M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; Chen, H.S.; et al. SEER Cancer Statistics Review 1975–2016; National Cancer Institute: Bethesda, MD, USA, 2018.
- Misiakos, P.E.; Karidis, N.P.; Kouraklis, G. Current treatment for colorectal liver metastases. World J. Gastroenterol. 2011, 17, 4067–4075. [Google Scholar] [CrossRef] [PubMed]
- Kow, A.W.C. Hepatic metastasis from colorectal cancer. J. Gastrointest. Oncol. 2019, 10, 1274–1298. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The Role of MicroRNAs in Colorectal Cancer. Cancer J. 2012, 18, 244–252. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Álvarez-Castro, A.; López-López, R.; Iglesias-Canle, J.; Suárez-Cunqueiro, M.M.; Muinelo-Romay, L. Circulating microRNAs as Promising Biomarkers in Colorectal Cancer. Cancers 2019, 11, 898. [Google Scholar] [CrossRef] [PubMed]
- Nasr, R.; Hammoud, M.S.; Nassar, F.; Mukherji, D.; Shamseddine, A.I.; Temraz, S. Inflammatory Markers and MicroRNAs: The Backstage Actors Influencing Prognosis in Colorectal Cancer Patients. Int. J. Mol. Sci. 2018, 19, 1867. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Marcuello, M.; Duran-Sanchon, S.; Moreno, L.; Lozano, J.J.; Bujanda, L.; Castells, A.; Gironella, M. Analysis of A 6-Mirna Signature in Serum from Colorectal Cancer Screening Participants as Non-Invasive Biomarkers for Advanced Adenoma and Colorectal Cancer Detection. Cancers 2019, 11, 1542. [Google Scholar] [CrossRef]
- Yamada, A.; Horimatsu, T.; Okugawa, Y.; Nishida, N.; Honjo, H.; Ida, H.; Kou, T.; Kusaka, T.; Sasaki, Y.; Yagi, M.; et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin. Cancer Res. 2015, 21, 4234–4242. [Google Scholar] [CrossRef]
- Ramzy, I.; Hasaballah, M.; Marzaban, R.; Shaker, O.; Soliman, Z.A. Evaluation of microRNAs-29a, 92a and 145 in colorectal carcinoma as candidate diagnostic markers: An Egyptian pilot study. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 508–515. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–665. [Google Scholar] [CrossRef]
- Wang, W.; Qu, A.; Liu, W.; Liu, Y.; Zheng, G.; Du, L.; Zhang, X.; Yang, Y.; Wang, C.; Chen, X. Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur. J. Cancer Care 2016, 26, e12448. [Google Scholar] [CrossRef]
- Yong, F.L.; Law, C.W.; Wang, C.W. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 2013, 13, 280. [Google Scholar] [CrossRef]
- Lv, Z.-C.; Fan, Y.-S.; Chen, H.-B.; Zhao, D.-W. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumor Biol. 2015, 36, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Baker, K.; Redman, M.W.; Wang, L.; Adams, S.V.; Yu, M.; Dickinson, B.; Makar, K.; Ulrich, N.; Böhm, J.; et al. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br. J. Cancer 2017, 117, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, N.; Wang, X.; Ren, H.; Wang, W.; Wang, S.; Song, Y.; Liu, Y.; Li, Y.; Zhou, X.; et al. Circulating serum microRNA-345 correlates with unfavorable pathological response to preoperative chemoradiotherapy in locally advanced rectal cancer. Oncotarget 2016, 7, 64233–64243. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-H.; Tsao, C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Liming, Z.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell. Physiol. Biochem. 2017, 43, 945–958. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.K.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2007, 27, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Farouk, S.; Khairy, A.; Salem, A.M.; Soliman, A.F.; El Din, N.G.B. Differential Expression of miR-21, miR-23a, and miR-27a, and Their Diagnostic Significance in Egyptian Colorectal Cancer Patients. Genet. Test. Mol. Biomark. 2020, 24, 825–834. [Google Scholar] [CrossRef]
- Kanaan, Z.; Rai, S.N.; Eichenberger, M.R.; Roberts, H.; Keskey, B.; Pan, J.; Galandiuk, S. Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann. Surg. 2012, 256, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Sabry, D.; El-Deek, S.E.M.; Maher, M.; El-Baz, M.A.H.; El-Bader, H.M.; Amer, E.; Hassan, E.A.; Fathy, W.; El-Deek, H.E.M. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol. Cell. Biochem. 2019, 454, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, D.; Guan, S.; Dong, M. Diagnostic role of circulating MiR-21 in colorectal cancer: A update meta-analysis. Ann. Med. 2021, 53, 87–102. [Google Scholar] [CrossRef]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A Current Overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar] [CrossRef]
- Di, Z.; Di, M.; Fu, W.; Tang, Q.; Liu, Y.; Lei, P.; Gu, X.; Liu, T.; Sun, M. Integrated Analysis Identifies a Nine-microRNA Signature Biomarker for Diagnosis and Prognosis in Colorectal Cancer. Front. Genet. 2020, 11, 192. [Google Scholar] [CrossRef]
- Li, G.; Wang, Q.; Li, Z.; Shen, Y. Serum miR-21 and miR-210 as promising non-invasive biomarkers for the diagnosis and prognosis of colorectal cancer. Rev. Esp. Enferm. Dig. 2020, 112, 832–837. [Google Scholar] [CrossRef]
- Chiang, Y.; Song, Y.; Wang, Z.; Chen, Y.; Yue, Z.; Xu, H.; Xing, C.; Liu, Z. Aberrant Expression of miR-203 and Its Clinical Significance in Gastric and Colorectal Cancers. J. Gastrointest. Surg. 2010, 15, 63–70. [Google Scholar] [CrossRef]
- Bovell, L.C.; Shanmugam, C.; Putcha, B.D.K.; Katkoori, V.R.; Zhang, B.; Bae, S.; Singh, K.P.; Grizzle, W.E.; Manne, U. The Prognostic Value of MicroRNAs Varies with Patient Race/Ethnicity and Stage of Colorectal Cancer. Clin. Cancer Res. 2013, 19, 3955–3965. [Google Scholar] [CrossRef]
- Deng, B.; Wang, B.; Fang, J.; Zhu, X.; Cao, Z.; Lin, Q.; Zhou, L.; Sun, X. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A. Sci. Rep. 2016, 6, 28301. [Google Scholar] [CrossRef] [PubMed]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.; Cogdell, D.; Calin, G.A.; Sun, B.; Kopetz, S.; Hamilton, S.R.; Zhang, W. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget 2016, 7, 11434–11449. [Google Scholar] [CrossRef]
- Huang, G.; Wei, B.; Chen, Z.; Wang, J.; Zhao, L.; Peng, X.; Liu, K.; Lai, Y.; Ni, L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark. Med. 2020, 14, 749–760. [Google Scholar] [CrossRef]
- Ye, H.; Hao, H.; Wang, J.; Chen, R.; Huang, Z. miR-203 as a novel biomarker for the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget Ther. 2017, 10, 3685–3696. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, S.-K.; Zhao, M.; Yang, M.; Zhong, J.-L.; Gu, Y.-Y.; Peng, H.; Che, Y.-Q.; Huang, C.-Z. Identification of a Circulating MicroRNA Signature for Colorectal Cancer Detection. PLoS ONE 2014, 9, e87451. [Google Scholar] [CrossRef]
- Yu, T.; Ma, P.; Wu, D.; Shu, Y.; Gao, W. Functions and mechanisms of microRNA-31 in human cancers. Biomed. Pharm. 2018, 108, 1162–1169. [Google Scholar] [CrossRef]
- Mi, B.; Li, Q.; Li, T.; Liu, G.; Sai, J. High miR-31-5p expression promotes colon adenocarcinoma progression by targeting TNS. Aging 2020, 12, 7480–7490. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Chen, Z.-H.; Chen, W. Novel circulating microRNAs expression profile in colon cancer: A pilot study. Eur. J. Med. Res. 2017, 22, 1–11. [Google Scholar] [CrossRef]
- Peng, X.; Wang, J.; Zhang, C.; Liu, K.; Zhao, L.; Chen, X.; Huang, G.; Lai, Y. A three-miRNA panel in serum as a noninvasive biomarker for colorectal cancer detection. Int. J. Biol. Markers 2020, 35, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-W.; Ming, X.-L.; Rong, Y.; Huang, C.-Q.; Weng, H.; Chen, H.; Bian, J.-M.; Wang, F.-B. Diagnostic Value Investigation and Bioinformatics Analysis of miR-31 in Patients with Lymph Node Metastasis of Colorectal Cancer. Anal. Cell. Pathol. 2019, 2019, 9740475. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, J.; Tong, Y.; Li, J.; Liu, B. miR-145-5p restrained cell growth, invasion, migration and tumorigenesis via modulating RHBDD1 in colorectal cancer via the EGFR-associated signaling pathway. Int. J. Biochem. Cell Biol. 2019, 117, 105641. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, L.; Ye, X.; Tao, M.; Wu, J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA. Pathol. Res. Pract. 2020, 216, 152872. [Google Scholar] [CrossRef]
- Maminezhad, H.; Ghanadian, S.; Pakravan, K.; Razmara, E.; Rouhollah, F.; Mossahebi-Mohammadi, M.; Babashah, S. A panel of six-circulating miRNA signature in serum and its potential diagnostic value in colorectal cancer. Life Sci. 2020, 258, 118226. [Google Scholar] [CrossRef]
- Luo, X.; Wu, Y.; Ji, M.; Zhang, S. Combined Plasma MicroRNA and Fecal Occult Blood Tests in Early Detection of Colorectal Cancer. Clin. Lab. 2019, 65, 65. [Google Scholar] [CrossRef]
- Eslamizadeh, S.; Heidari, M.; Agah, S.; Faghihloo, E.; Ghazi, H.; Mirzaei, A.; Akbari, A. The Role of MicroRNA Signature as Diagnostic Biomarkers in Different Clinical Stages of Colorectal Cancer. Cell J. 2018, 20, 220–230. [Google Scholar]
- Liu, Q.; Yang, W.; Luo, Y.; Hu, S.; Zhu, L. Correlation between miR-21 and miR-145 and the incidence and prognosis of colorectal cancer. JBUON 2018, 23, 29–35. [Google Scholar] [PubMed]
- Liu, N.; Jiang, F.; Han, X.-Y.; Li, M.; Chen, W.-J.; Liu, Q.-C.; Liao, C.-X.; Lv, Y.-F. MiRNA-155 promotes the invasion of colorectal cancer SW-480 cells through regulating the Wnt/β-catenin. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 101–109. [Google Scholar]
- Orosz, E.; Kiss, I.; Gyöngyi, Z.; Varjas, T. Expression of Circulating miR-155, miR-21, miR-221, miR-30a, miR-34a and miR-29a: Comparison of Colonic and Rectal Cancer. In Vivo 2018, 32, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-T.; Wang, J.-L.; Du, W.; Hong, J.; Zhao, S.-L.; Wang, Y.-C.; Xiong, H.; Chen, H.-M.; Fang, J.-Y. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis 2011, 32, 1207–1215. [Google Scholar] [CrossRef]
- Schou, J.; Rossi, S.; Jensen, B.V.; Nielsen, D.L.; Pfeiffer, P.; Høgdall, E.V.; Yilmaz, M.; Tejpar, S.; Delorenzi, M.; Kruhøffer, M.; et al. miR-345 in Metastatic Colorectal Cancer: A Non-Invasive Biomarker for Clinical Outcome in Non-KRAS Mutant Patients Treated with 3rd Line Cetuximab and Irinotecan. PLoS ONE 2014, 9, e99886. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, R.; Yang, F.; Cheng, R.; Chen, X.; Cui, S.; Gu, Y.; Sun, W.; You, C.; Liu, Z.; et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA. Mol. Cancer 2017, 16, 1–17. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, Y.; Gao, J.; Fu, J.; Liu, C.; Liu, Y.; Song, C.; Zhu, S.; Leng, Y.; Wang, G.; et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF. Br. J. Cancer 2014, 110, 450–458. [Google Scholar] [CrossRef]
- Deng, Y.H.; Deng, Z.H.; Hao, H.; Wu, X.L.; Gao, H.; Tang, S.H.; Tang, H. MicroRNA-23a promotes colorectal cancer cell survival by targeting PDK. Exp. Cell Res. 2018, 373, 171–179. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.-W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M.; et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 2016, 37, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Cellura, D.; Pickard, K.; Quaratino, S.; Parker, H.; Strefford, J.; Thomas, G.; Mitter, R.; Mirnezami, A.; Peake, N. miR-19–Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer. Mol. Cancer Res. 2015, 13, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, Z.; Lai, D.; Sun, J.; He, C.; Chu, Z.; Ye, H.; Chen, S.; Wang, J. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. Br. J. Cancer 2012, 107, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genom. 2013, 14, 589. [Google Scholar] [CrossRef]
- Ellermeier, C.; Vang, S.; Cleveland, K.; Durand, W.; Resnick, M.B.; Brodsky, A.S. Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res. 2014, 34, 3957–3967. [Google Scholar]
- Ahmed, E.K.; Fahmy, S.A.; Effat, H.; Wahab, A.H.A. Circulating miR-210 and miR-1246 as potential biomarkers for differentiating hepatocellular carcinoma from metastatic tumors in the liver. J. Med. Biochem. 2019, 38, 109–117. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.-M.; Zhang, Z.-J.; Li, X. miR-210 is a Serological Biomarker for Predicting Recurrence and Prognosis of Colon Carcinoma Patients with Liver Metastases After Radiofrequency Ablation Treatment. Cancer Manag. Res. 2020, 12, 9077–9085. [Google Scholar] [CrossRef]
- Drusco, A.; Nuovo, G.J.; Zanesi, N.; Di Leva, G.; Pichiorri, F.; Volinia, S.; Fernandez, C.; Antenucci, A.; Costinean, S.; Bottoni, A.; et al. MicroRNA Profiles Discriminate among Colon Cancer Metastasis. PLoS ONE 2014, 9, e96670. [Google Scholar] [CrossRef]
- Pecqueux, M.; Liebetrau, I.; Werft, W.; Dienemann, H.; Muley, T.; Pfannschmidt, J.; Müssle, B.; Rahbari, N.N.; Schölch, S.; Büchler, M.W.; et al. A Comprehensive MicroRNA Expression Profile of Liver and Lung Metastases of Colorectal Cancer with Their Corresponding Host Tissue and Its Prognostic Impact on Survival. Int. J. Mol. Sci. 2016, 17, 1755. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, T.; Zheng, X.; Yang, S.; Xu, K.; Chen, X.; Xu, F.; Wang, L.; Shen, Y.; Wang, T.; et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018, 39, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, M.; Iinuma, H.; Yagi, T.; Matsuda, K.; Hashiguchi, Y. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology 2017, 92, 360–370. [Google Scholar] [CrossRef]
- Yin, J.; Bai, Z.; Song, J.; Yang, Y.; Wang, J.; Han, W.; Zhang, J.; Meng, H.; Ma, X.; Yang, Y.; et al. Differential expression of serum miR-126, miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer. Chin. J. Cancer Res. 2014, 26, 95–103. [Google Scholar] [PubMed]
- Nassar, F.J.; El Sabban, M.; Zgheib, N.K.; Tfayli, A.; Boulos, F.; Jabbour, M.; Saghir, N.S.E.L.; Talhouk, R.; Bazarbachi, A.; Calin, G.A.; et al. miRNA as Potential Biomarkers of Breast Cancer in the Lebanese Population and in Young Women: A Pilot Study. PLoS ONE 2014, 9, e107566. [Google Scholar] [CrossRef] [PubMed]
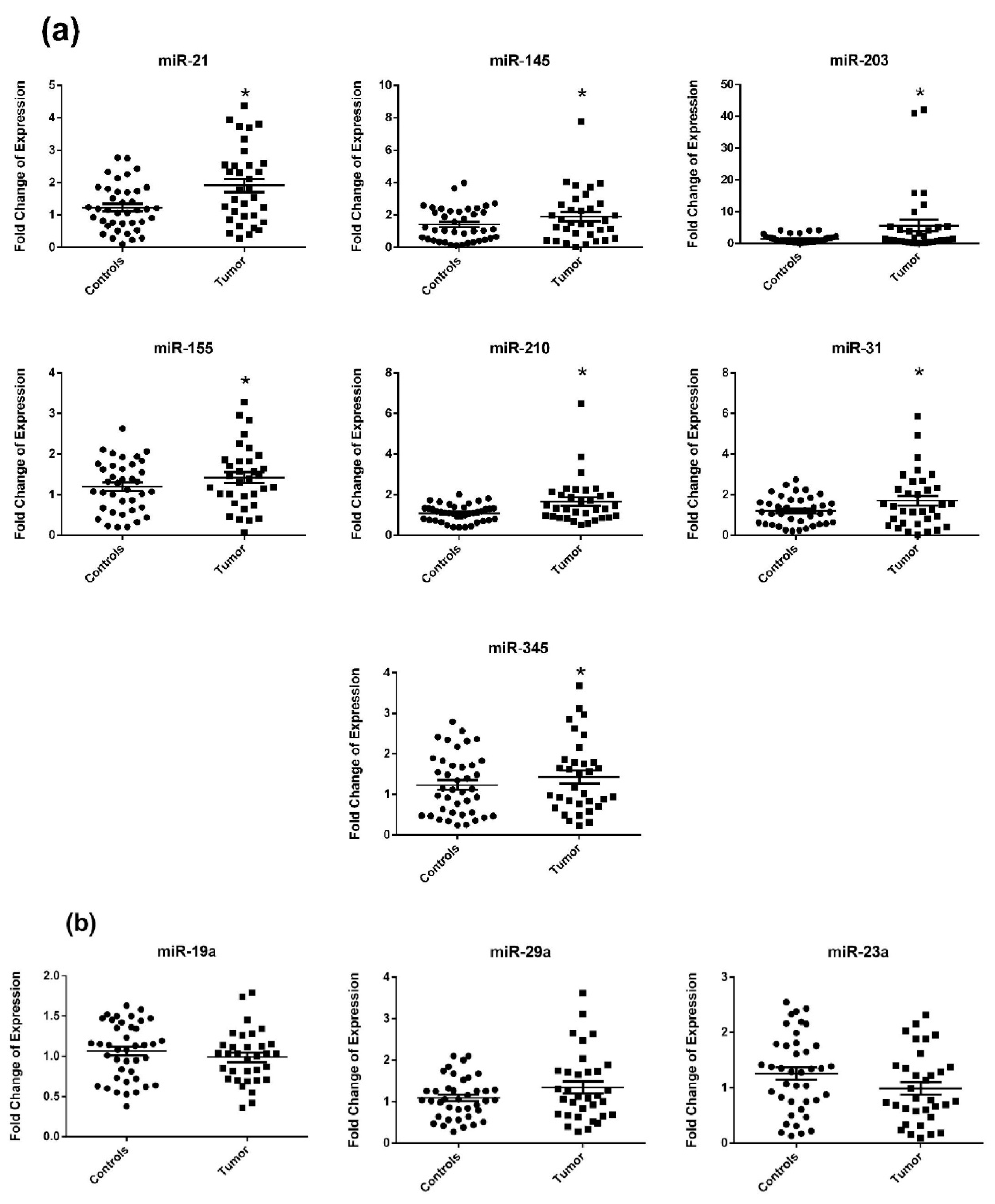


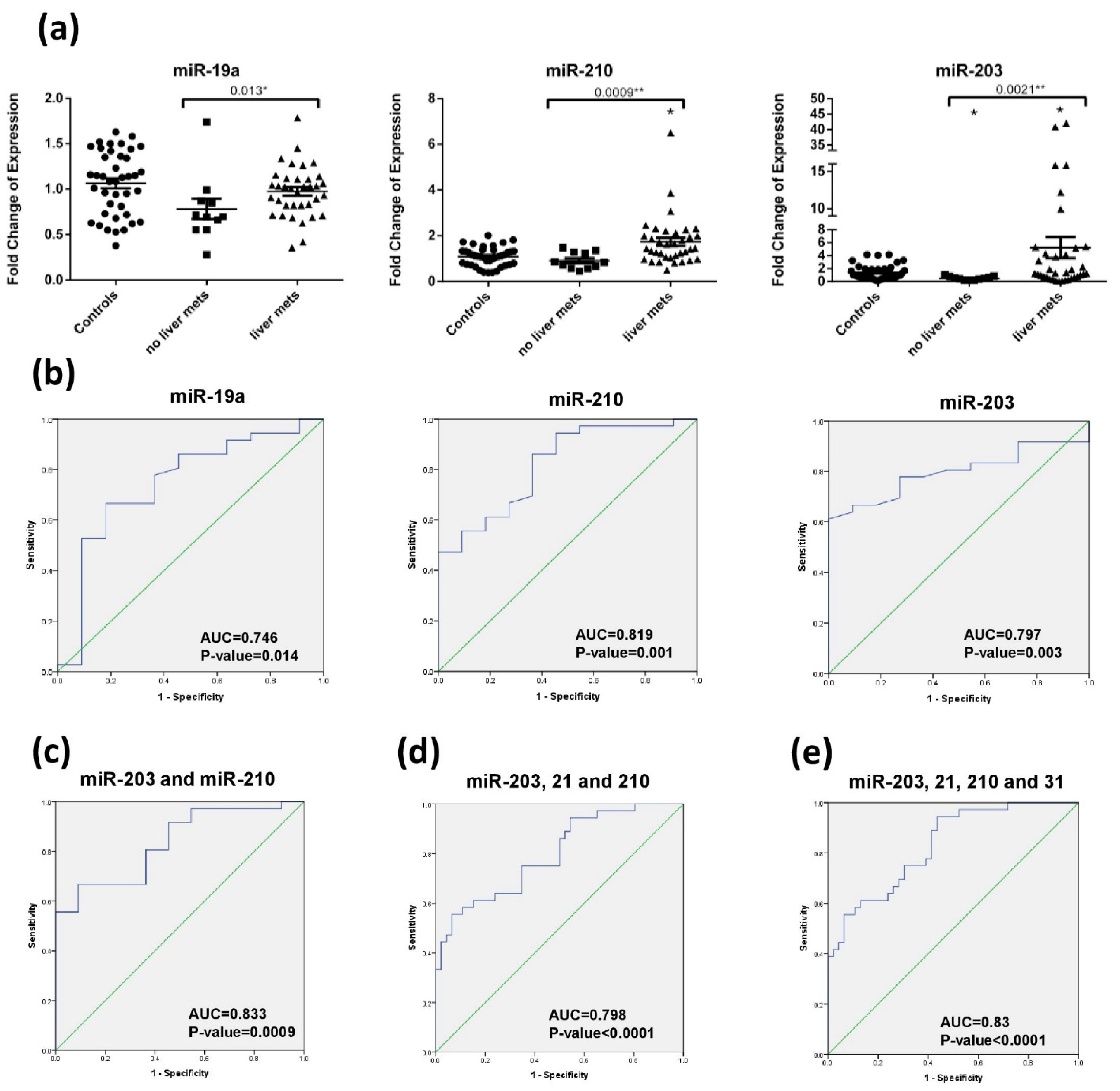
| Variable | All CRC Patients | Surgery Naïve | Postoperative | Recurrent | Healthy Subjects |
|---|---|---|---|---|---|
| Sample | 62 (58.5) | 33 (53.2) | 15 (24.2) | 14 (22.6) | 44 (41.5) |
| Age (years) | 58 ± 13.49 | 56.3 ± 2.2 | 56.26 ± 13.86 | 63.9 ± 14.25 | 41.72 ± 8.97 |
| BMI (kg/m2) | 27.17 ± 6.34 | 27.6 ± 6.37 | 25.1 ± 7.66 | 28.37 ± 4.35 | 26.39 ± 0.39 |
| Gender | |||||
| Male | 38 (61.3) | 21 (63.6) | 6 (40) | 11 (78.6) | 18 (40.9) |
| Female | 24 (38.7) | 12 (36.4) | 9 (60) | 3 (21.4) | 26 (59.1) |
| Nationality | |||||
| Lebanese | 50 (82) | 27 (81.8) | 12 (85.7) | 11 (78.6) | 44 (100) |
| Iraqi | 8 (13.1) | 5 (15.2) | 1 (7.1) | 2 (14.3) | |
| Other | 3 (4.9) | 1 (3) | 1 (7.1) | 1 (7.1) | |
| Smoking habits | |||||
| No | 47 (75.8) | 26 (78.8) | 11 (73.3) | 11 (78.6) | 27 (61.4) |
| Yes | 15 (24.2) | 7 (21.2) | 4 (26.7) | 3 (21.4) | 17 (38.6) |
| Alcohol intake | |||||
| No | 48 (77.4) | 26 (78.8) | 11 (73.3) | 11 (78.6) | 34 (81) |
| Yes | 14 (22.6) | 7 (21.2) | 4 (26.7) | 3 (21.4) | 8 (19) |
| Family Hx of Cancer | |||||
| No | 35 (57.4) | 19 (57.6) | 7 (46.7) | 9 (64.3) | |
| Yes | 26 (42.6) | 14 (42.4) | 8 (53.3) | 4 (30.8) | |
| Tumor Sidedness | |||||
| Left | 36 (60) | 24 (72.7) | 6 (42.9) | 6 (42.9) | |
| Right | 15 (25) | 3 (9.1) | 7 (50) | 5 (35.7) | |
| Rectum | 9 (15) | 5 (15.2) | 1 (7.1) | 3 (21.4) | |
| Liver Mets | |||||
| No | 15 (24.2) | 5 (15.2) | 4 (26.7) | 6 (42.9) | |
| Yes | 47 (75.8) | 28 (84.8) | 11 (73.3) | 8 (57.1) | |
| KRAS mutation | |||||
| No | 26 (48.1) | 16 (53.3) | 5 (33.3) | 6 (50) | |
| Yes | 28 (51.9) | 14 (46.7) | 8 (53.3) | 6 (50) | |
| NRAS mutation | |||||
| No | 46 (93.9) | 28 (93.3) | 9 (100) | 10 (90.9) | |
| Yes | 3 (6.1) | 2 (6.7) | 0 | 1 (9.1) | |
| BRAF mutation | |||||
| No | 49 (100) | 28 (100) | 9 (100) | 11 (100) | |
| Yes | 0 | 0 | 0 |
| miRNA | AUC | SE | p-Value | 95% CI | Youden’s Index | Cut-Off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | DA (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-210 | 0.682 | 0.055 | 0.003 | 0.574–0.79 | 0.288 | 10.46 | 44.7 | 84.1 | 75 | 56 | 62 |
| miR-21 | 0.651 | 0.057 | 0.013 | 0.539–0.76 | 0.247 | 2.81 | 38.3 | 86.4 | 75 | 53 | 59 |
| miR-210+21 | 0.731 | 0.052 | 0.0001 | 0.63–0.832 | 0.35 | 0.464 | 87.2 | 47.7 | 65 | 79 | 69 |
| miRNA | AUC | SE | p-Value | 95% CI | Youden’s Index | Cut-Off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | DA (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-19a | 0.746 | 0.091 | 0.014 | 0.569–0.924 | 0.485 | 6.025 | 66.7 | 81.8 | 93 | 45 | 72 |
| miR-210 | 0.819 | 0.069 | 0.001 | 0.684–0.955 | 0.497 | 11.115 | 86.1 | 63.6 | 89 | 58 | 81 |
| miR-203 | 0.797 | 0.063 | 0.003 | 0.673–0.92 | 0.611 | 15.65 | 61.1 | 100 | 100 | 44 | 70 |
| miR-210+203 | 0.833 | 0.063 | 0.0009 | 0.711–0.956 | 0.576 | 0.838 | 66.7 | 90.9 | 96 | 45 | 72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; Helou, R.E.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R.; et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics 2021, 11, 341. https://doi.org/10.3390/diagnostics11020341
Nassar FJ, Msheik ZS, Itani MM, Helou RE, Hadla R, Kreidieh F, Bejjany R, Mukherji D, Shamseddine A, Nasr RR, et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics. 2021; 11(2):341. https://doi.org/10.3390/diagnostics11020341
Chicago/Turabian StyleNassar, Farah J., Zahraa S. Msheik, Maha M. Itani, Remie El Helou, Ruba Hadla, Firas Kreidieh, Rachelle Bejjany, Deborah Mukherji, Ali Shamseddine, Rihab R. Nasr, and et al. 2021. "Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis" Diagnostics 11, no. 2: 341. https://doi.org/10.3390/diagnostics11020341
APA StyleNassar, F. J., Msheik, Z. S., Itani, M. M., Helou, R. E., Hadla, R., Kreidieh, F., Bejjany, R., Mukherji, D., Shamseddine, A., Nasr, R. R., & Temraz, S. N. (2021). Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics, 11(2), 341. https://doi.org/10.3390/diagnostics11020341









