Urinary Cell-Free DNA in Bladder Cancer Detection
Abstract
1. Introduction
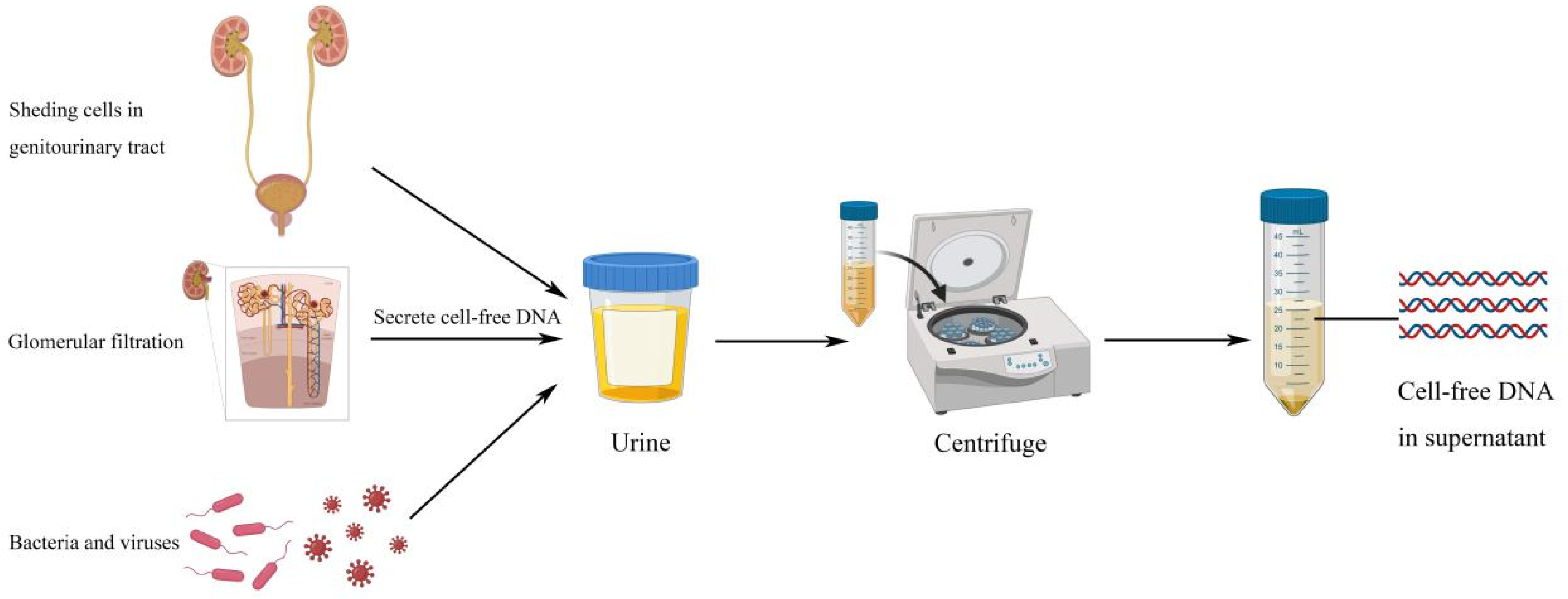
2. Urinary Cell-Free DNA Features
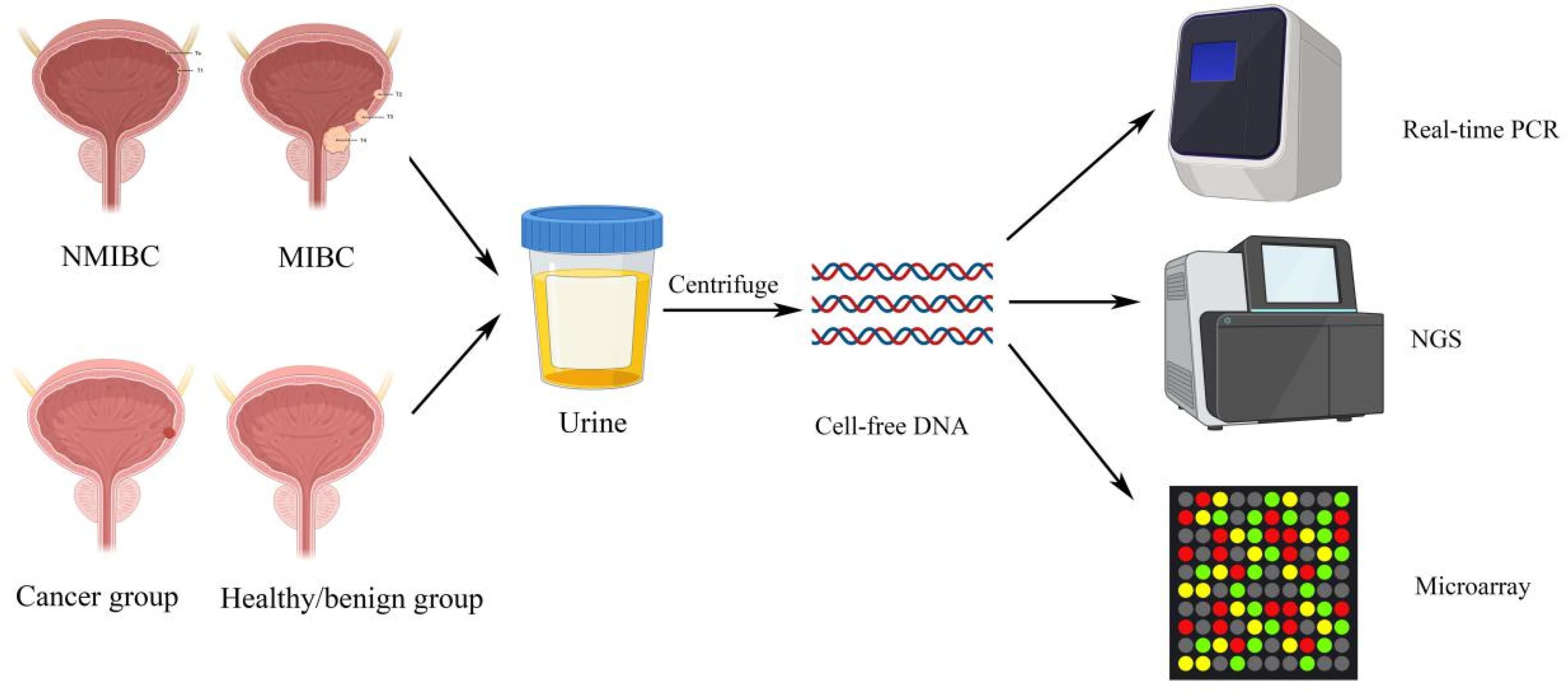
3. Bladder Cancers Detection and Diagnosis
3.1. Urinary Cell-free DNA Integrity
3.2. Urinary Cell-free DNA Concentrations
3.3. Urinary Cell-free DNA Sequencing
3.4. Urinary Cell-free DNA Expression
4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Teoh, J.Y.; Huang, J.; Ko, W.Y.; Lok, V.; Choi, P.; Ng, C.F.; Sengupta, S.; Mostafid, H.; Kamat, A.M.; Black, P.C.; et al. Global Trends of Bladder Cancer Incidence and Mortality, and Their Associations with Tobacco Use and Gross Domestic Product Per Capita. Eur. Urol. 2020, 78, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Tsui, K.H.; Shen, L.C.; Huang, H.W.; Wang, S.N.; Chang, P.L. Urinary cell-free DNA as a potential tumor marker for bladder cancer. Int. J. Biol. Markers 2007, 22, 287–294. [Google Scholar] [CrossRef]
- Ward, D.G.; Bryan, R.T. Liquid biopsies for bladder cancer. Transl. Androl. Urol. 2017, 6, 331–335. [Google Scholar] [CrossRef]
- Ou, Z.; Li, K.; Yang, T.; Dai, Y.; Chandra, M.; Ning, J.; Wang, Y.; Xu, R.; Gao, T.; Xie, Y.; et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin. Transl. Med. 2020, 9, 4. [Google Scholar] [CrossRef]
- Salvi, S.; Casadio, V. Urinary Cell-Free DNA: Potential and Applications. Methods Mol. Biol. 2019, 1909, 201–209. [Google Scholar] [CrossRef]
- Lu, T.; Li, J. Clinical applications of urinary cell-free DNA in cancer: Current insights and promising future. Am. J. Cancer Res. 2017, 7, 2318–2332. [Google Scholar] [PubMed]
- Bryzgunova, O.E.; Laktionov, P.P. Extracellular Nucleic Acids in Urine: Sources, Structure, Diagnostic Potential. Acta Nat. 2015, 7, 48–54. [Google Scholar] [CrossRef]
- Brisuda, A.; Pazourkova, E.; Soukup, V.; Horinek, A.; Hrbacek, J.; Capoun, O.; Svobodova, I.; Pospisilova, S.; Korabecna, M.; Mares, J.; et al. Urinary Cell-Free DNA Quantification as Non-Invasive Biomarker in Patients with Bladder Cancer. Urol. Int. 2016, 96, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Zancan, M.; Galdi, F.; Di Tonno, F.; Mazzariol, C.; Orlando, C.; Malentacchi, F.; Agostini, M.; Maran, M.; Del Bianco, P.; Fabricio, A.S.; et al. Evaluation of cell-free DNA in urine as a marker for bladder cancer diagnosis. Int. J. Biol. Markers 2009, 24, 147–155. [Google Scholar] [CrossRef]
- Casadio, V.; Calistri, D.; Tebaldi, M.; Bravaccini, S.; Gunelli, R.; Martorana, G.; Bertaccini, A.; Serra, L.; Scarpi, E.; Amadori, D.; et al. Urine cell-free DNA integrity as a marker for early bladder cancer diagnosis: Preliminary data. Urol. Oncol. 2013, 31, 1744–1750. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Conte, D.; Leon, M.; Ciricione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003, 21, 3902–3908. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Casadio, V.; Salvi, S. Urinary Cell-Free DNA: Isolation, Quantification, and Quality Assessment. Methods Mol. Biol. 2019, 1909, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Tabach, Y.; Kogan-Sakin, I.; Buganim, Y.; Solomon, H.; Goldfinger, N.; Hovland, R.; Ke, X.S.; Oyan, A.M.; Kalland, K.H.; Rotter, V.; et al. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS ONE 2011, 6, e14632. [Google Scholar] [CrossRef]
- Nord, H.; Segersten, U.; Sandgren, J.; Wester, K.; Busch, C.; Menzel, U.; Komorowski, J.; Dumanski, J.P.; Malmstrom, P.U.; Diaz de Stahl, T. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. Int. J. Cancer 2010, 126, 1390–1402. [Google Scholar] [CrossRef]
- Hansel, D.E.; Swain, E.; Dreicer, R.; Tubbs, R.R. HER2 overexpression and amplification in urothelial carcinoma of the bladder is associated with MYC coamplification in a subset of cases. Am. J. Clin. Pathol. 2008, 130, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Erhola, M.; Toyokuni, S.; Okada, K.; Tanaka, T.; Hiai, H.; Ochi, H.; Uchida, K.; Osawa, T.; Nieminen, M.M.; Alho, H.; et al. Biomarker evidence of DNA oxidation in lung cancer patients: Association of urinary 8-hydroxy-2’-deoxyguanosine excretion with radiotherapy, chemotherapy, and response to treatment. FEBS Lett. 1997, 409, 287–291. [Google Scholar] [CrossRef]
- Zhang, J.; Tong, K.L.; Li, P.K.; Chan, A.Y.; Yeung, C.K.; Pang, C.C.; Wong, T.Y.; Lee, K.C.; Lo, Y.M. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clin. Chem. 1999, 45, 1741–1746. [Google Scholar] [CrossRef]
- Dyer, A.R.; Greenland, P.; Elliott, P.; Daviglus, M.L.; Claeys, G.; Kesteloot, H.; Ueshima, H.; Stamler, J.; Group, I.R. Evaluation of measures of urinary albumin excretion in epidemiologic studies. Am. J. Epidemiol. 2004, 160, 1122–1131. [Google Scholar] [CrossRef]
- Zancan, M.; Franceschini, R.; Mimmo, C.; Vianello, M.; Di Tonno, F.; Mazzariol, C.; Malossini, G.; Gion, M. Free DNA in urine: A new marker for bladder cancer? Preliminary data. Int. J. Biol. Markers 2005, 20, 134–136. [Google Scholar] [CrossRef]
- Cheng, T.H.T.; Jiang, P.; Teoh, J.Y.C.; Heung, M.M.S.; Tam, J.C.W.; Sun, X.; Lee, W.S.; Ni, M.; Chan, R.C.K.; Ng, C.F.; et al. Noninvasive Detection of Bladder Cancer by Shallow-Depth Genome-Wide Bisulfite Sequencing of Urinary Cell-Free DNA for Methylation and Copy Number Profiling. Clin. Chem. 2019, 65, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Birkenkamp-Demtroder, K.; Nordentoft, I.; Christensen, E.; Hoyer, S.; Reinert, T.; Vang, S.; Borre, M.; Agerbaek, M.; Jensen, J.B.; Orntoft, T.F.; et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur. Urol. 2016, 70, 75–82. [Google Scholar] [CrossRef]
- Sylvester, R.J.; van der Meijden, A.P.; Oosterlinck, W.; Witjes, J.A.; Bouffioux, C.; Denis, L.; Newling, D.W.; Kurth, K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur. Urol. 2006, 49, 466–477, discussion 475–477. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Sasano, H.; Yamaki, H.; Shizawa, S.; Kikuchi, A.; Shineha, R.; Akaishi, T.; Satomi, S.; Nagura, H. Topoisomerase II alpha expression in esophageal squamous cell carcinoma. Anticancer Res. 1999, 19, 1873–1880. [Google Scholar]
- Kim, Y.H.; Yan, C.; Lee, I.S.; Piao, X.M.; Byun, Y.J.; Jeong, P.; Kim, W.T.; Yun, S.J.; Kim, W.J. Value of urinary topoisomerase-IIA cell-free DNA for diagnosis of bladder cancer. Investig. Clin. Urol. 2016, 57, 106–112. [Google Scholar] [CrossRef]
- Dwivedi, U.S.; Kumar, A.; Das, S.K.; Trivedi, S.; Kumar, M.; Sunder, S.; Singh, P.B. Relook TURBT in superficial bladder cancer: Its importance and its correlation with the tumor ploidy. Urol. Oncol. 2009, 27, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Kuzaka, B.; Janiak, M.; Wlodarski, K.H.; Radziszewski, P.; Wlodarski, P.K. Expression of bone morphogenetic protein-2 and -7 in urinary bladder cancer predicts time to tumor recurrence. Arch. Med. Sci. 2015, 11, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Laatio, L.; Myllynen, P.; Serpi, R.; Rysa, J.; Ilves, M.; Lappi-Blanco, E.; Ruskoaho, H.; Vahakangas, K.; Puistola, U. BMP-4 expression has prognostic significance in advanced serous ovarian carcinoma and is affected by cisplatin in OVCAR-3 cells. Tumour Biol. 2011, 32, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Hassan, M.K.; Pattnaik, N.; Mohapatra, N.; Dixit, M. Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers. PLoS ONE 2017, 12, e0186977. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Kim, Y.H.; Jeong, P.; Piao, X.M.; Byun, Y.J.; Seo, S.P.; Kang, H.W.; Kim, W.T.; Lee, J.Y.; Ryu, D.H.; et al. Urinary Cell-Free DNA IQGAP3/BMP4 Ratio as a Prognostic Marker for Non-Muscle-Invasive Bladder Cancer. Clin. Genitourin. Cancer 2019, 17, e704–e711. [Google Scholar] [CrossRef]
- Cheng, T.H.; Jiang, P.; Tam, J.C.; Sun, X.; Lee, W.S.; Yu, S.C.; Teoh, J.Y.; Chiu, P.K.; Ng, C.F.; Chow, K.M.; et al. Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Utting, M.; Werner, W.; Dahse, R.; Schubert, J.; Junker, K. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: A minimally invasive method for the detection of bladder cancer. Clin. Cancer Res. 2002, 8, 35–40. [Google Scholar]
| Parameters/Techniques | Markers | Descriptions |
|---|---|---|
| ucfDNA integrity | Long ucfDNA fragments (>250 bp) | Long fragmented ucfDNA, originated from necrotic tumor cells, were significantly more abundant in bladder cancer patients than in healthy individuals and symptomatic non-cancerous patients. |
| ucfDNA concentration | Total amount of ucfDNA | Significantly higher total amount of ucfDNA in bladder cancer patients than in healthy and benign patients. Total amount of ucfDNA was higher in patients with higher grades and stages of bladder cancer. |
| 400-bp ucfDNA/urine creatinine (Ucr) and PicoGreen ucfDNA/UCr | ucfDNA isolated from both methods were significantly higher in bladder cancer patients. 400-bp ucfDNA/UCr was more sensitive and specific than PicoGreen ucfDNA/UCr. | |
| ucfDNA 250 ng/mL concentration threshold | All cancer patients with concentrations of ucfDNA exceeded this threshold, while only less than half of the healthy controls did. | |
| ucfDNA sequencing | TERT, FGFR3, TP53, PIK3CA and KRAS 5-genes panel | The 5-gene panel was generated from frequently mutated genes in bladder cancers. Highest total number of mutations were observed in bladder cancer patients. It had a high sensitivity in detection and monitoring of bladder cancers, and this panel had an area under curve (AUC) of 0.94. |
| Methylation deconvolution, global methylation and copy number alterations (CNAs) | High concordance between tumor and ucfDNA in terms of hypomethylation and CNAs. Bladder cancer patients exhibited significantly hypomethylation and CNAs. Post-operative patients showed lower levels of all three parameters. Sensitivity and specificity were higher when all three parameters were combined. | |
| Personalized assay | Tumor-specific ucfDNA isolated from personalized assay were higher in progressive/metastatic disease than in recurrent non-muscle invasive bladder cancer (NMIBC). Genomic variants in ucfDNA could be detected prior to tumor resection. Chemotherapy could affect levels of tumor-specific ucfDNA. | |
| ucfDNA Expression | TopoIIA ucfDNA | TopoIIA ucfDNA expression level was significantly higher in bladder cancer patients than inhealthy individuals, and higher in muscle invasive bladder cancer (MIBC) than in NMIBC. |
| IQGAP3/BMP4 and IQGAP3/FAM107A | IQGAP3/BMP4 was overexpressed in high grade and stage bladder cancers, IQGAP3/FAM107A was overexpressed in larger tumor size and progression. Both higher ratios were associated with worse progression-free survival (PFS), and a high IQGAP3/BMP4 ratio was also associated with worse recurrence-free survival (RFS). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tse, R.T.-H.; Zhao, H.; Wong, C.Y.-P.; Cheng, C.K.-L.; Kong, A.W.-Y.; Peng, Q.; Chiu, P.K.-F.; Ng, C.-F.; Teoh, J.Y.-C. Urinary Cell-Free DNA in Bladder Cancer Detection. Diagnostics 2021, 11, 306. https://doi.org/10.3390/diagnostics11020306
Tse RT-H, Zhao H, Wong CY-P, Cheng CK-L, Kong AW-Y, Peng Q, Chiu PK-F, Ng C-F, Teoh JY-C. Urinary Cell-Free DNA in Bladder Cancer Detection. Diagnostics. 2021; 11(2):306. https://doi.org/10.3390/diagnostics11020306
Chicago/Turabian StyleTse, Ryan Tsz-Hei, Hongda Zhao, Christine Yim-Ping Wong, Carol Ka-Lo Cheng, Angel Wing-Yan Kong, Qiang Peng, Peter Ka-Fung Chiu, Chi-Fai Ng, and Jeremy Yuen-Chun Teoh. 2021. "Urinary Cell-Free DNA in Bladder Cancer Detection" Diagnostics 11, no. 2: 306. https://doi.org/10.3390/diagnostics11020306
APA StyleTse, R. T.-H., Zhao, H., Wong, C. Y.-P., Cheng, C. K.-L., Kong, A. W.-Y., Peng, Q., Chiu, P. K.-F., Ng, C.-F., & Teoh, J. Y.-C. (2021). Urinary Cell-Free DNA in Bladder Cancer Detection. Diagnostics, 11(2), 306. https://doi.org/10.3390/diagnostics11020306







