Clinico-Pathological Importance of miR-146a in Lung Cancer
Abstract
1. Introduction
2. miR-146a Expression and Regulation
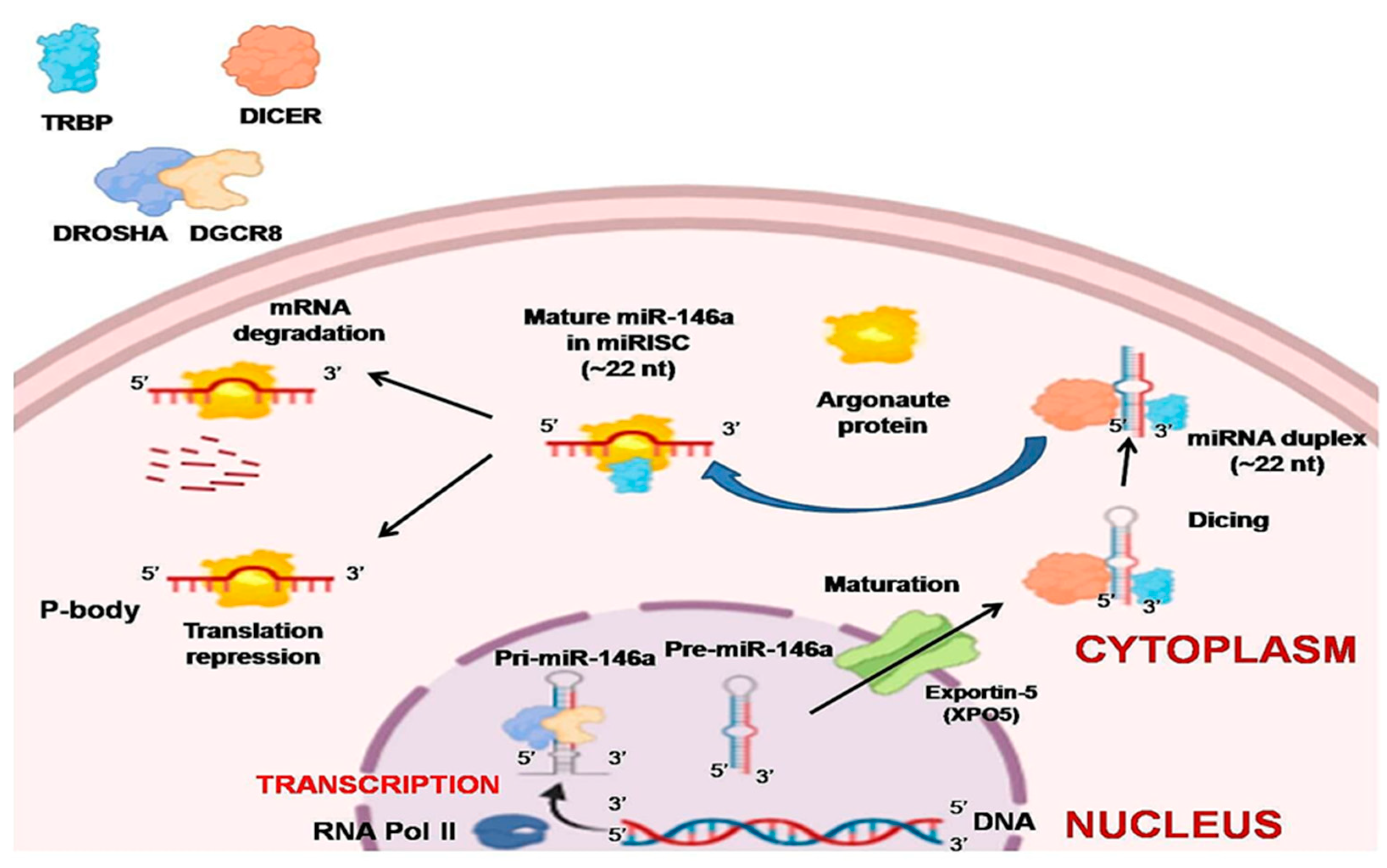
2.1. Biogenesis
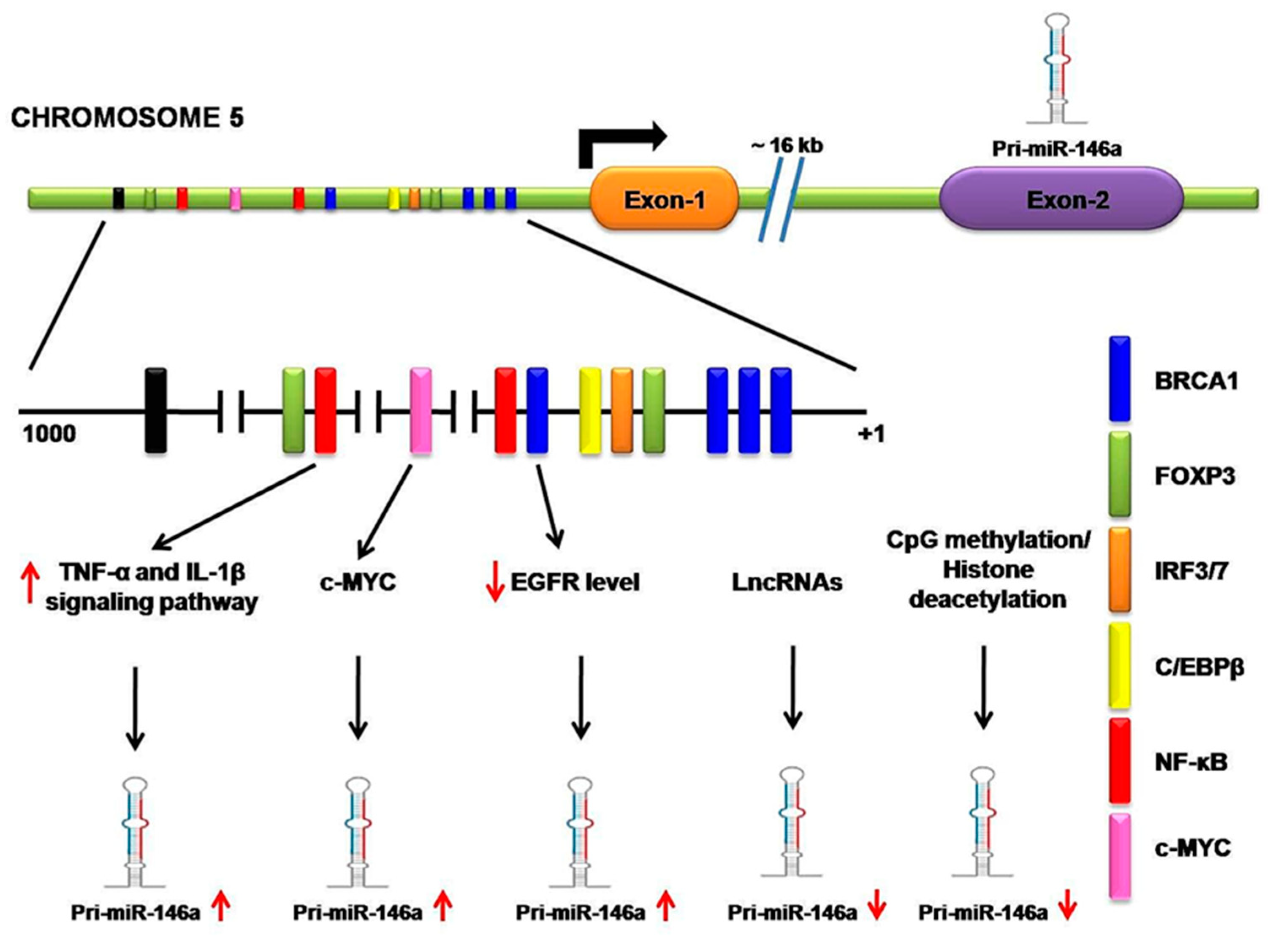
2.2. Transcriptional and Post-Transcriptional Regulation
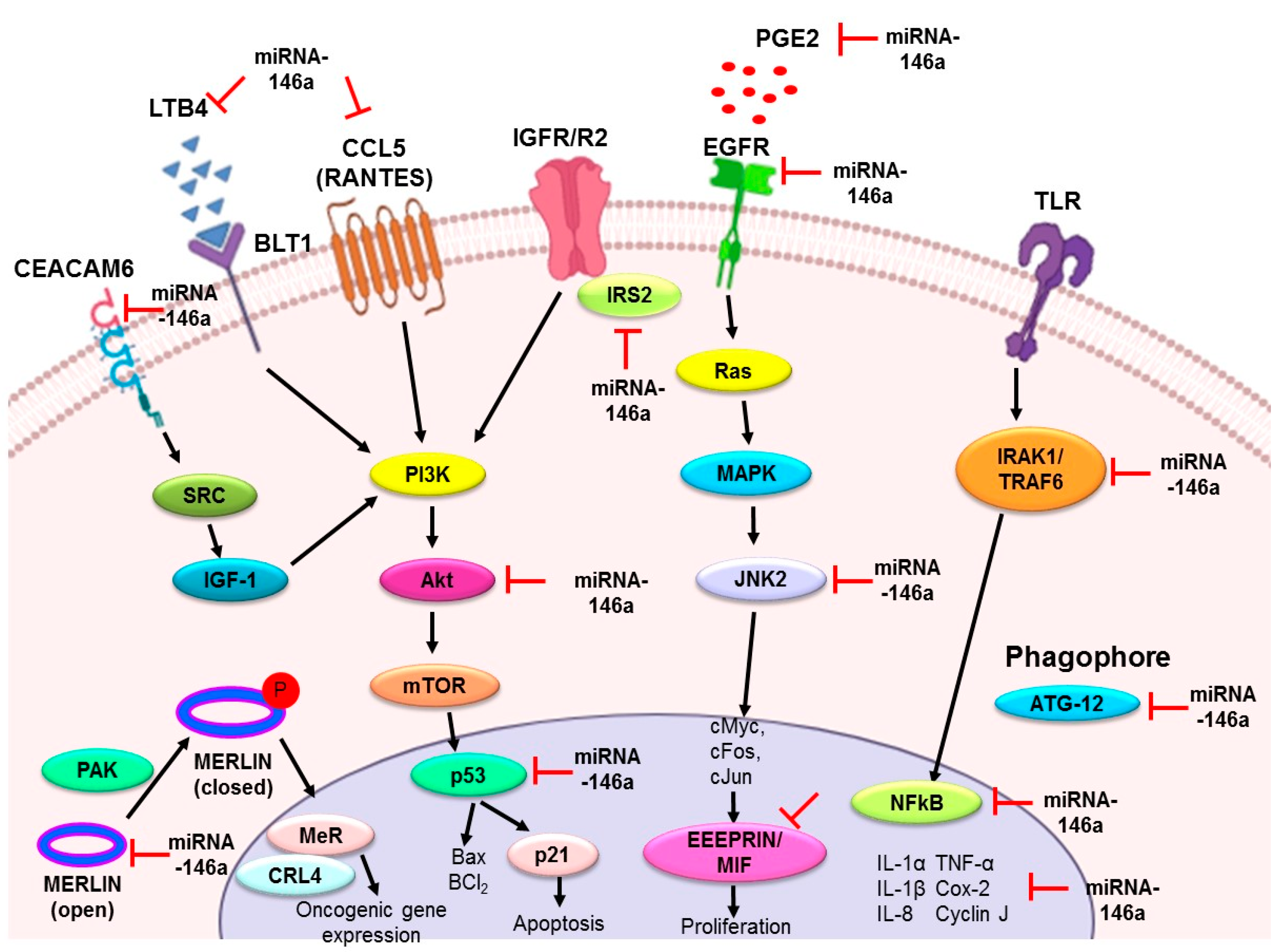
3. Role of miR-146a in Lung Cancer
3.1. miR-146a as an Antiproliferative and Proapoptotic Agent
3.2. miR-146a as an Anti-Inflammatory Agent
3.3. miR-146a as a Metastatic Modulator
4. miR-146a as a Biomarker in Lung Cancer
4.1. Diagnostic Potential
4.2. Prognostic Potential
5. Potential of miRNA-146a in Lung Cancer
5.1. miR-146a Restoration as an Adjunct Therapy to Current Therapeutic Agents
5.2. Potential for Small Molecule-Upregulation of miR-146a
5.3. Future Perspectives and Conclusion
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Zheng, D.; Haddadin, S.; Wang, Y.; Gu, L.Q.; Perry, M.C.; Freter, C.E.; Wang, M.X. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 2011, 4, 575–586. [Google Scholar] [PubMed]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Pammolli, F.; Magazzini, L.; Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011, 10, 428–438. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic Assay Based on hsa-miR-205 Expression Distinguishes Squamous From Nonsquamous Non–Small-Cell Lung Carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef]
- Landi, M.T.; Zhao, Y.; Rotunno, M.; Koshiol, J.; Liu, H.; Bergen, A.W.; Rubagotti, M.; Goldstein, A.M.; Linnoila, I.; Marincola, F.M.; et al. MicroRNA Expression Differentiates Histology and Predicts Survival of Lung Cancer. Clin. Cancer Res. 2010, 16, 430–441. [Google Scholar] [CrossRef]
- Boeri, M.; Verri, C.; Conte, D.; Roz, L.; Modena, P.; Facchinetti, F.; Calabrò, E.; Croce, C.M.; Pastorino, U.; Sozzi, G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 3713–3718. [Google Scholar] [CrossRef] [PubMed]
- Gilad, S.; Lithwick-Yanai, G.; Barshack, I.; Benjamin, S.; Krivitsky, I.; Edmonston, T.B.; Bibbo, M.; Thurm, C.; Horowitz, L.; Huang, Y.; et al. Classification of the Four Main Types of Lung Cancer Using a MicroRNA-Based Diagnostic Assay. J. Mol. Diagn. 2012, 14, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zhong, Y.; Wu, L.; Yang, D.; Ye, S.; Zhang, M. Prognostic value of microRNAs in lung cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2018, 10, 67–77. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A MicroRNA Regulator with a Macro Role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Yin, J.; Zhao, J.; Hu, W.; Yang, G.; Yu, H.; Wang, R.; Wang, L.; Zhang, G.; Fu, W.; Dai, L.; et al. Disturbance of the let-7/LIN28 double-negative feedback loop is associated with radio- and chemo-resistance in non-small cell lung cancer. PLoS ONE 2017, 12, e0172787. [Google Scholar] [CrossRef]
- Lagos-Quintana, M. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Chen, G.; Umelo, I.A.; Lv, S.; Teugels, E.; Fostier, K.; Kronenberger, P.; Dewaele, A.; Sadones, J.; Geers, C.; De Grève, J. miR-146a Inhibits Cell Growth, Cell Migration and Induces Apoptosis in Non-Small Cell Lung Cancer Cells. PLoS ONE 2013, 8, e60317. [Google Scholar] [CrossRef] [PubMed]
- Iacona, J.R.; Monteleone, N.J.; Lutz, C.S. miR-146a suppresses 5-lipoxygenase activating protein (FLAP) expression and Leukotriene B4 production in lung cancer cells. Oncotarget 2018, 9, 26751–26769. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yi, R.; Cullen, B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2004, 24, 138–148. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.-J.; Park, J.-E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef]
- Burke, J.M.; Kelenis, D.P.; Kincaid, R.P.; Sullivan, C.S. A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA. RNA 2014, 20, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yeo, J.; Lee, J.H.; Cho, J.; Seo, D.; Kim, J.-S.; Kim, V.N. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell Rep. 2014, 9, 1061–1074. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-Body Formation Is a Consequence, Not the Cause, of RNA-Mediated Gene Silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Cornett, A.L.; Lutz, C.S. Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA 2014, 20, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Yu, D.; Lee, Y.-S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2007, 40, 43–50. [Google Scholar] [CrossRef]
- Forloni, M.; Dogra, S.K.; Dong, Y.; Conte, D.; Ou, J.; Zhu, L.J.; Deng, A.; Mahalingam, M.; Green, M.R.; Wajapeyee, N. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. eLife 2014, 3, e01460. [Google Scholar] [CrossRef]
- Kulis, M.; Esteller, M. DNA Methylation and Cancer. In Epigenetics and Cancer, Part A; Elsevier: Amsterdam, The Netherlands, 2010; pp. 27–56. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Sun, X.X.; Ma, X.; Chen, Z.N. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol. Cancer 2015, 14, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Gao, H.; Ren, L.; Gu, J.; Zhang, Y.; Zhang, Y. Demethylation of the miR-146a promoter by 5-Aza-2’-deoxycytidine correlates with delayed progression of castration-resistant prostate cancer. BMC Cancer 2014, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- de Giorgio, A.; Krell, J.; Harding, V.; Stebbing, J.; Castellano, L. Emerging Roles of Competing Endogenous RNAs in Cancer: Insights from the Regulation of PTEN. Mol. Cell. Biol. 2013, 33, 3976–3982. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-X.; Zhao, W.; Mao, L.-W.; Wang, Y.-L.; Xia, L.-Q.; Cao, M.; Shen, J.; Chen, J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018, 104, 25–33. [Google Scholar] [CrossRef]
- Han, W.; Du, X.; Liu, M.; Wang, J.; Sun, L.; Li, Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int. J. Biol. Macromol. 2019, 121, 270–278. [Google Scholar] [CrossRef]
- Kumaraswamy, E.; Wendt, K.L.; Augustine, L.A.; Stecklein, S.R.; Sibala, E.C.; Li, D.; Gunewardena, S.; Jensen, R.A. BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene 2014, 34, 4333–4346. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, C.; Chen, D.; Yang, W.-H.; Liu, X.; Liu, C.-G.; Dugas, C.M.; Tang, F.; Zheng, P.; Liu, Y.; et al. FOXP3 Controls an miR-146/NF-κB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer Res. 2015, 75, 1703–1713. [Google Scholar] [CrossRef]
- Sun, M.; Fang, S.; Li, W.; Li, C.; Wang, L.; Wang, F.; Wang, Y. Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark. 2015, 15, 33–40. [Google Scholar] [CrossRef]
- Larner-Svensson, H.M.; Williams, A.E.; Tsitsiou, E.; Perry, M.M.; Jiang, X.; Chung, K.F.; Lindsay, M.A. Pharmacological studies of the mechanism and function of interleukin-1β-induced miRNA-146a expression in primary human airway smooth muscle. Respir. Res. 2010, 11, 1–13. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-W.; Gao, L.; Dang, Y.-W.; Li, P.; Li, Z.-Y.; Chen, G.; Luo, D.-Z. Protective potential of miR-146a-5p and its underlying molecular mechanism in diverse cancers: A comprehensive meta-analysis and bioinformatics analysis. Cancer Cell Int. 2019, 19, 1–21. [Google Scholar] [CrossRef]
- Pavel, A.B.; Campbell, J.D.; Liu, G.; Elashoff, D.; Dubinett, S.; Smith, K.; Whitney, D.; Lenburg, M.E.; Spira, A. Alterations in Bronchial Airway miRNA Expression for Lung Cancer Detection. Cancer Prev. Res. 2017, 10, 651–659. [Google Scholar] [CrossRef]
- Scagliotti, G.V.; Selvaggi, G.; Novello, S.; Hirsch, F.R. The Biology of Epidermal Growth Factor Receptor in Lung Cancer. Clin. Cancer Res. 2004, 10, 4227s–4232s. [Google Scholar] [CrossRef]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Li, Y.; Liu, X.; Jafari, F.A.; Zhang, X.; Sun, Q.; Ma, Z. Cryptotanshinone Suppresses Non-Small Cell Lung Cancer via microRNA-146a-5p/EGFR Axis. Int. J. Biol. Sci. 2019, 15, 1072–1079. [Google Scholar] [CrossRef]
- Zucker, S.; Hymowitz, M.; Rollo, E.E.; Mann, R.; Conner, C.E.; Cao, J.; Foda, H.D.; Tompkins, D.C.; Toole, B.P. Tumorigenic Potential of Extracellular Matrix Metalloproteinase Inducer. Am. J. Pathol. 2001, 158, 1921–1928. [Google Scholar] [CrossRef]
- Huang, W.T.; He, R.Q.; Li, X.J.; Ma, J.; Peng, Z.G.; Zhong, J.C.; Hu, X.H.; Chen, G. miR-146a-5p targets TCSF and influences cell growth and apoptosis to repress NSCLC progression. Oncol. Rep. 2019, 41, 2226–2240. [Google Scholar] [CrossRef]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, T.; Wilson, J.; Kulbe, H.; Li, N.F.; Leinster, D.A.; Charles, K.; Klemm, F.; Pukrop, T.; Binder, C.; Balkwill, F.R. Macrophages Induce Invasiveness of Epithelial Cancer Cells Via NF-κB and JNK. J. Immunol. 2005, 175, 1197–1205. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Xie, F.; Peng, J.; Wu, X. Macrophage migration inhibitory factor promotes Warburg effect via activation of the NF-κB/HIF-1α pathway in lung cancer. Int. J. Mol. Med. 2017, 41, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-M.; Liu, J.-C. Effect and molecular mechanism of mir-146a on proliferation of lung cancer cells by targeting and regulating MIF gene. Asian Pac. J. Trop. Med. 2016, 9, 806–811. [Google Scholar] [CrossRef]
- Li, Y.-L.; Wang, J.; Zhang, C.-Y.; Shen, Y.-Q.; Wang, H.-M.; Ding, L.; Gu, Y.-C.; Lou, J.-T.; Zhao, X.-T.; Ma, Z.-L.; et al. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget 2016, 7, 59287–59298. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Xu, Z.; Wu, G.; Chen, X.; Huang, Y.; Wang, Y.; Jiang, W.; Ke, B. Up-regulation of miR-146a increases the sensitivity of non-small cell lung cancer to DDP by downregulating cyclin J. BMC Cancer 2017, 17, 1–14. [Google Scholar] [CrossRef]
- Park, D.H.; Jeon, H.S.; Lee, S.Y.; Choi, Y.Y.; Lee, H.W.; Yoon, S.; Lee, J.C.; Yoon, Y.S.; Kim, D.S.; Na, M.J.; et al. MicroRNA-146a inhibits epithelial mesenchymal transition in non-small cell lung cancer by targeting insulin receptor substrate 2. Int. J. Oncol. 2015, 47, 1545–1553. [Google Scholar] [CrossRef]
- Heuberger, J.; Birchmeier, W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a002915. [Google Scholar] [CrossRef]
- Tan, W.; Liao, Y.; Qiu, Y.; Liu, H.; Tan, D.; Wu, T.; Tang, M.; Zhang, S.; Wang, H. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP). Cancer Lett. 2018, 428, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, E.I.; Meza-Sosa, K.F.; López-Sevilla, Y.; Camacho-Concha, N.; Sánchez, N.C.; Pérez-Martínez, L.; Pedraza-Alva, G. Merlin negative regulation by miR-146a promotes cell transformation. Biochem. Biophys. Res. Commun. 2015, 468, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.; Charles, K.A.; Mantovani, A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 2005, 7, 211–217. [Google Scholar] [CrossRef]
- Gomes, M.; Teixeira, A.L.; Coelho, A.; Araújo, A.; Medeiros, R. The Role of Inflammation in Lung Cancer. In Advances in Experimental Medicine and Biology; Springer Basel: Basel, Switzerland, 2014; pp. 1–23. [Google Scholar] [CrossRef]
- Richardson, C.M.; Sharma, R.A.; Cox, G.; O’Byrne, K.J. Epidermal growth factor receptors and cyclooxygenase-2 in the pathogenesis of non-small cell lung cancer: Potential targets for chemoprevention and systemic therapy. Lung Cancer 2003, 39, 1–13. [Google Scholar] [CrossRef]
- Saba, R.; Sorensen, D.L.; Booth, S.A. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef]
- Perry, M.M.; Moschos, S.A.; Williams, A.E.; Shepherd, N.J.; Larner-Svensson, H.M.; Lindsay, M.A. Rapid Changes in MicroRNA-146a Expression Negatively Regulate the IL-1β-Induced Inflammatory Response in Human Lung Alveolar Epithelial Cells. J. Immunol. 2008, 180, 5689–5698. [Google Scholar] [CrossRef] [PubMed]
- Lambert, K.A.; Roff, A.N.; Panganiban, R.P.; Douglas, S.; Ishmael, F.T. MicroRNA-146a is induced by inflammatory stimuli in airway epithelial cells and augments the anti-inflammatory effects of glucocorticoids. PLoS ONE 2018, 13, e0205434. [Google Scholar] [CrossRef]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Orjalo, A.V.; Rodier, F.; Lithgow, G.J.; Campisi, J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging 2009, 1, 402–411. [Google Scholar] [CrossRef]
- Bui, N.; Woodward, B.; Johnson, A.; Husain, H. Novel Treatment Strategies for Brain Metastases in Non-small-cell Lung Cancer. Curr. Treat. Opt. Oncol. 2016, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.G.; Sanders, A.J.; Katoh, M.; Ungefroren, H.; Gieseler, F.; Prince, M.; Thompson, S.K.; Zollo, M.; Spano, D.; Dhawan, P.; et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin. Cancer Biol. 2015, 35, S244–S275. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, E.C.; Chuaqui, R.F.; Liotta, L.A. General mechanisms of metastasis. Cancer 1997, 80, 1529–1537. [Google Scholar] [CrossRef]
- Cheung, K.J.; Ewald, A.J. A collective route to metastasis: Seeding by tumor cell clusters. Science 2016, 352, 167–169. [Google Scholar] [CrossRef]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell. Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef]
- Shahriar, A.; Ghaleh-Aziz Shiva, G.; Ghader, B.; Farhad, J.; Hosein, A.; Parsa, H. The dual role of mir-146a in metastasis and disease progression. Biomed. Pharm. 2020, 126, 110099. [Google Scholar] [CrossRef]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef]
- Bleau, A.M.; Redrado, M.; Nistal-Villan, E.; Villalba, M.; Exposito, F.; Redin, E.; de Aberasturi, A.L.; Larzabal, L.; Freire, J.; Gomez-Roman, J.; et al. miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 2018, 414, 257–267. [Google Scholar] [CrossRef]
- Ghuwalewala, S.; Ghatak, D.; Das, S.; Das, P.; Butti, R.; Gorain, M.; Kundu, G.C.; Roychoudhury, S. MiR-146a-dependent regulation of CD24/AKT/β-catenin axis drives cancer stem cell phenotype in oral squamous cell carcinoma. bioRxiv 2019, 429068. [Google Scholar] [CrossRef]
- Saunders, N.A.; Simpson, F.; Thompson, E.W.; Hill, M.M.; Endo-Munoz, L.; Leggatt, G.; Minchin, R.F.; Guminski, A. Role of intratumoural heterogeneity in cancer drug resistance: Molecular and clinical perspectives. Embo Mol. Med. 2012, 4, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, N.C.; Medrano-Jimenez, E.; Aguilar-Leon, D.; Perez-Martinez, L.; Pedraza-Alva, G. Tumor Necrosis Factor-Induced miR-146a Upregulation Promotes Human Lung Adenocarcinoma Metastasis by Targeting Merlin. DNA Cell Biol. 2020, 39, 484–497. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Wang, R.J.; Zheng, Y.H.; Wang, P.; Zhang, J.Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 765–771. [Google Scholar] [PubMed]
- Mohamed, R.H.; Pasha, H.F.; Gad, D.M.; Toam, M.M. miR-146a and miR-196a-2 genes polymorphisms and its circulating levels in lung cancer patients. J. Biochem. 2019, 166, 323–329. [Google Scholar] [CrossRef]
- Sodhi, K.K.; Bahl, C.; Singh, N.; Behera, D.; Sharma, S. Functional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol. 2015, 11, 2159–2173. [Google Scholar] [CrossRef]
- Yuwen, D.L.; Sheng, B.B.; Liu, J.; Wenyu, W.; Shu, Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharm. Sci 2017, 21, 2650–2658. [Google Scholar]
- Wu, C.; Cao, Y.; He, Z.; He, J.; Hu, C.; Duan, H.; Jiang, J. Serum Levels of miR-19b and miR-146a as Prognostic Biomarkers for Non-Small Cell Lung Cancer. Tohoku J. Exp. Med. 2014, 232, 85–95. [Google Scholar] [CrossRef]
- Yoon, K.-A.; Yoon, H.; Park, S.; Jang, H.-J.; Zo, J.I.; Lee, H.-S.; Lee, J.S. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non–small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2012, 144, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.-G.; Zhou, X.-M.; Cui, Z.-G.; Hou, G. Effects of common polymorphisms in miR-146a and miR-196a2 on lung cancer susceptibility: A meta-analysis. J. Thorac. Dis. 2016, 8, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Ma, X.; Sun, C.; Chen, L. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem. Biol. Interact. 2010, 184, 431–438. [Google Scholar] [CrossRef]
- Jiang, P.; Jia, W.; Wei, X.; Zhang, X.; Wang, C.; Li, B.; Song, T.; Yang, J.; Zhu, D.; Meng, Y. MicroRNA-146a regulates cisplatin-resistance of non-small cell lung cancer cells by targeting NF-kappaB pathway. Int. J. Clin. Exp. Pathol. 2017, 10, 11545–11553. [Google Scholar]
- Pang, L.; Lu, J.; Huang, J.; Xu, C.; Li, H.; Yuan, G.; Cheng, X.; Chen, J. Upregulation of miR-146a increases cisplatin sensitivity of the non-small cell lung cancer A549 cell line by targeting JNK-2. Oncol. Lett. 2017, 14, 7745–7752. [Google Scholar] [CrossRef][Green Version]
- Beauchemin, N.; Arabzadeh, A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013, 32, 643–671. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, H.; Li, Y.; Yuan, Y.; Chen, B.; Sun, S. Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Sci. 2020, 111, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Li, Y.; Sun, R.; Yuan, Y.; Sun, S.; Zhang, Y. CEACAM6 promotes cisplatin resistance in lung adenocarcinoma and is regulated by microRNA-146a and microRNA-26a. Thorac. Cancer 2020, 11, 2473–2482. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wan, C.; Zhang, W.; Guan, L.; Tian, G.; Zhang, F.; Ding, W. MiR-146a regulates PM1 -induced inflammation via NF-kappaB signaling pathway in BEAS-2B cells. Environ. Toxicol. 2018, 33, 743–751. [Google Scholar] [CrossRef]
- Mehta, M.; Tewari, D.; Gupta, G.; Awasthi, R.; Singh, H.; Pandey, P.; Chellappan, D.K.; Wadhwa, R.; Collet, T.; Hansbro, P.M.; et al. Oligonucleotide therapy: An emerging focus area for drug delivery in chronic inflammatory respiratory diseases. Chem. Biol. Interact. 2019, 308, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor MicroRNA-34. Cancer Res. 2010, 70, 5923–5930. [Google Scholar] [CrossRef]
- Stahlhut, C.; Slack, F.J. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle 2015, 14, 2171–2180. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Yeung, M.L.; Starost, M.F.; Hosmane, R.S.; Jeang, K.-T. Identification of Small Molecules That Suppress MicroRNA Function and Reverse Tumorigenesis. J. Biol. Chem. 2010, 285, 24707–24716. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, D.A.; Garner, A.L. Approaches for the Discovery of Small Molecule Ligands Targeting microRNAs. In Topics in Medicinal Chemistry; Springer International Publishing: New York City, NY, USA, 2017; pp. 79–110. [Google Scholar] [CrossRef]
- Velagapudi, S.P.; Vummidi, B.R.; Disney, M.D. Small molecule chemical probes of microRNA function. Curr. Opin. Chem. Biol. 2015, 24, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, W.; Zhou, M.; Kooger, R.; Zhang, Y. Small Molecules Modulating Biogenesis or Processing of microRNAs with Therapeutic Potentials. Curr. Med. Chem. 2013, 20, 3604–3612. [Google Scholar] [CrossRef]
- Deiters, A. Small Molecule Modifiers of the microRNA and RNA Interference Pathway. AAPS J. 2009, 12, 51–60. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Jan, C.-I.; Peng, C.-Y.; Lai, Y.-C.; Hu, F.-W.; Yu, C.-C. Activation of microRNA-494-targeting Bmi1 and ADAM10 by silibinin ablates cancer stemness and predicts favourable prognostic value in head and neck squamous cell carcinomas. Oncotarget 2015, 6, 24002–24016. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Rodova, M.; Nanta, R.; Meeker, D.; Van Veldhuizen, P.J.; Srivastava, R.K.; Shankar, S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro-oncology 2013, 15, 691–706. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Y.; Xie, C.; Yin, X.; Liu, Y.; Cao, Y.; Fang, Y.; Lin, X.; Xu, Y.; Xu, W.; et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int. J. Cancer 2014, 135, 1286–1296. [Google Scholar] [CrossRef]
- Zaman, M.S.; Chen, Y.; Deng, G.; Shahryari, V.; Suh, S.O.; Saini, S.; Majid, S.; Liu, J.; Khatri, G.; Tanaka, Y.; et al. The functional significance of microRNA-145 in prostate cancer. Br. J. Cancer 2010, 103, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.-H.; Hung, J.-H.; Chang, C.-W.; Weng, Y.-T.; Wu, M.-J.; Chen, P.-S. Solasodine inhibits invasion of human lung cancer cell through downregulation of miR-21 and MMPs expression. Chem. Biol. Interact. 2017, 268, 129–135. [Google Scholar] [CrossRef]
- Wei, Y.; Zou, Z.; Becker, N.; Anderson, M.; Sumpter, R.; Xiao, G.; Kinch, L.; Koduru, P.; Christudass, C.S.; Veltri, R.W.; et al. EGFR-Mediated Beclin 1 Phosphorylation in Autophagy Suppression, Tumor Progression, and Tumor Chemoresistance. Cell 2013, 154, 1269–1284. [Google Scholar] [CrossRef]
- Qu, J.; Chen, X.; Sun, Y.-Z.; Zhao, Y.; Cai, S.-B.; Ming, Z.; You, Z.-H.; Li, J.-Q. In Silico Prediction of Small Molecule-miRNA Associations Based on the HeteSim Algorithm. Mol. Ther. Nucleic Acids 2019, 14, 274–286. [Google Scholar] [CrossRef]
- Wang, C.-C.; Chen, X.; Qu, J.; Sun, Y.-Z.; Li, J.-Q. RFSMMA: A New Computational Model to Identify and Prioritize Potential Small Molecule–MiRNA Associations. J. Chem. Inf. Model. 2019, 59, 1668–1679. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, D.; Cuk, K.; Burwinkel, B.; Yang, R. Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front. Genet. 2013, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1–17. [Google Scholar] [CrossRef]
- Zhong, H.; Lu, J.; Jing, S.; Xi, J.; Yan, C.; Song, J.; Luo, S.; Zhao, C. Low-dose rituximab lowers serum Exosomal miR-150-5p in AChR-positive refractory myasthenia gravis patients. J. Neuroimmunol. 2020, 348, 577383. [Google Scholar] [CrossRef]
- Corral-Fernandez, N.E.; Salgado-Bustamante, M.; Martinez-Leija, M.E.; Cortez-Espinosa, N.; Garcia-Hernandez, M.H.; Reynaga-Hernandez, E.; Quezada-Calvillo, R.; Portales-Perez, D.P. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Stuckrath, I.; Rack, B.; Janni, W.; Jager, B.; Pantel, K.; Schwarzenbach, H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 2015, 6, 13387–13401. [Google Scholar] [CrossRef] [PubMed]
- Vang, S.; Wu, H.T.; Fischer, A.; Miller, D.H.; MacLaughlan, S.; Douglass, E.; Comisar, L.; Steinhoff, M.; Collins, C.; Smith, P.J.; et al. Identification of ovarian cancer metastatic miRNAs. PLoS ONE 2013, 8, e58226. [Google Scholar]
| Molecular Target | Clinical Significance | References | |
|---|---|---|---|
| miR-146a | COX2 | Decreases basal prostaglandins | [35] |
| Cyclin D1, Cyclin D2 | Discourages cell proliferation | [63] | |
| IRS2 | Discourages EMT/cancer progression | [65] | |
| IL-6, IL-8, RANTES | Prevents cytokine overproduction | [73,101] | |
| IRAK1, TRAF6, NF-kB | Weakens TNFα inflammatory stimuli and promotes cisplatin sensitivity | [72,101] | |
| Cyclin J | Promotes cisplatin chemosensitivity | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wani, J.A.; Majid, S.; Khan, A.; Arafah, A.; Ahmad, A.; Jan, B.L.; Shah, N.N.; Kazi, M.; Rehman, M.U. Clinico-Pathological Importance of miR-146a in Lung Cancer. Diagnostics 2021, 11, 274. https://doi.org/10.3390/diagnostics11020274
Wani JA, Majid S, Khan A, Arafah A, Ahmad A, Jan BL, Shah NN, Kazi M, Rehman MU. Clinico-Pathological Importance of miR-146a in Lung Cancer. Diagnostics. 2021; 11(2):274. https://doi.org/10.3390/diagnostics11020274
Chicago/Turabian StyleWani, Javaid Ahmad, Sabhiya Majid, Andleeb Khan, Azher Arafah, Ajaz Ahmad, Basit Latief Jan, Naveed Nazir Shah, Mohsin Kazi, and Muneeb U. Rehman. 2021. "Clinico-Pathological Importance of miR-146a in Lung Cancer" Diagnostics 11, no. 2: 274. https://doi.org/10.3390/diagnostics11020274
APA StyleWani, J. A., Majid, S., Khan, A., Arafah, A., Ahmad, A., Jan, B. L., Shah, N. N., Kazi, M., & Rehman, M. U. (2021). Clinico-Pathological Importance of miR-146a in Lung Cancer. Diagnostics, 11(2), 274. https://doi.org/10.3390/diagnostics11020274








