Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Sampling Strategy and miRNA Analysis
2.2.1. Sample Collection and RNA Extraction from the Serum
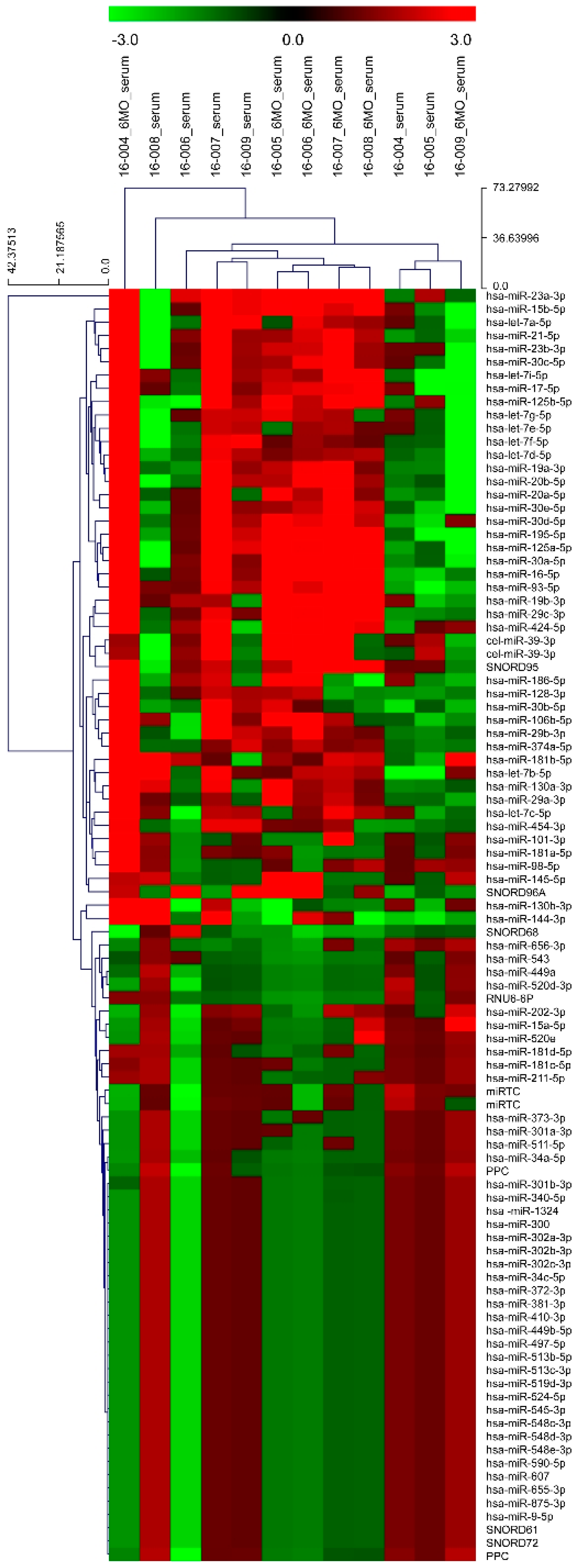
2.2.2. Profiling by miRNA Array Analysis
2.2.3. Validation by Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.2.4. Chemicals
2.2.5. Data Analysis and Statistical Methods
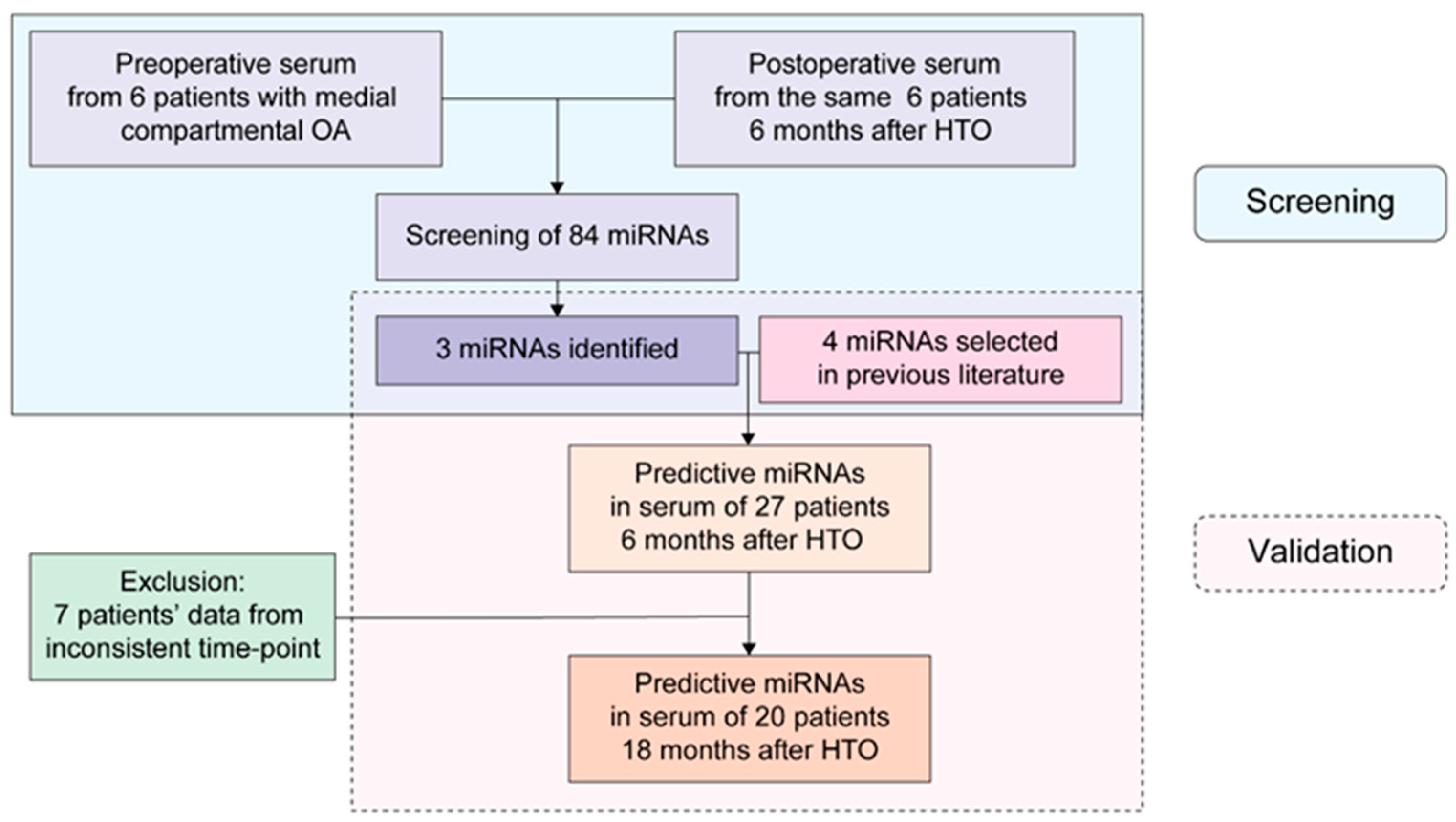
3. Results
3.1. Identification of Differentially Expressed miRNAs in the Profiling Stage
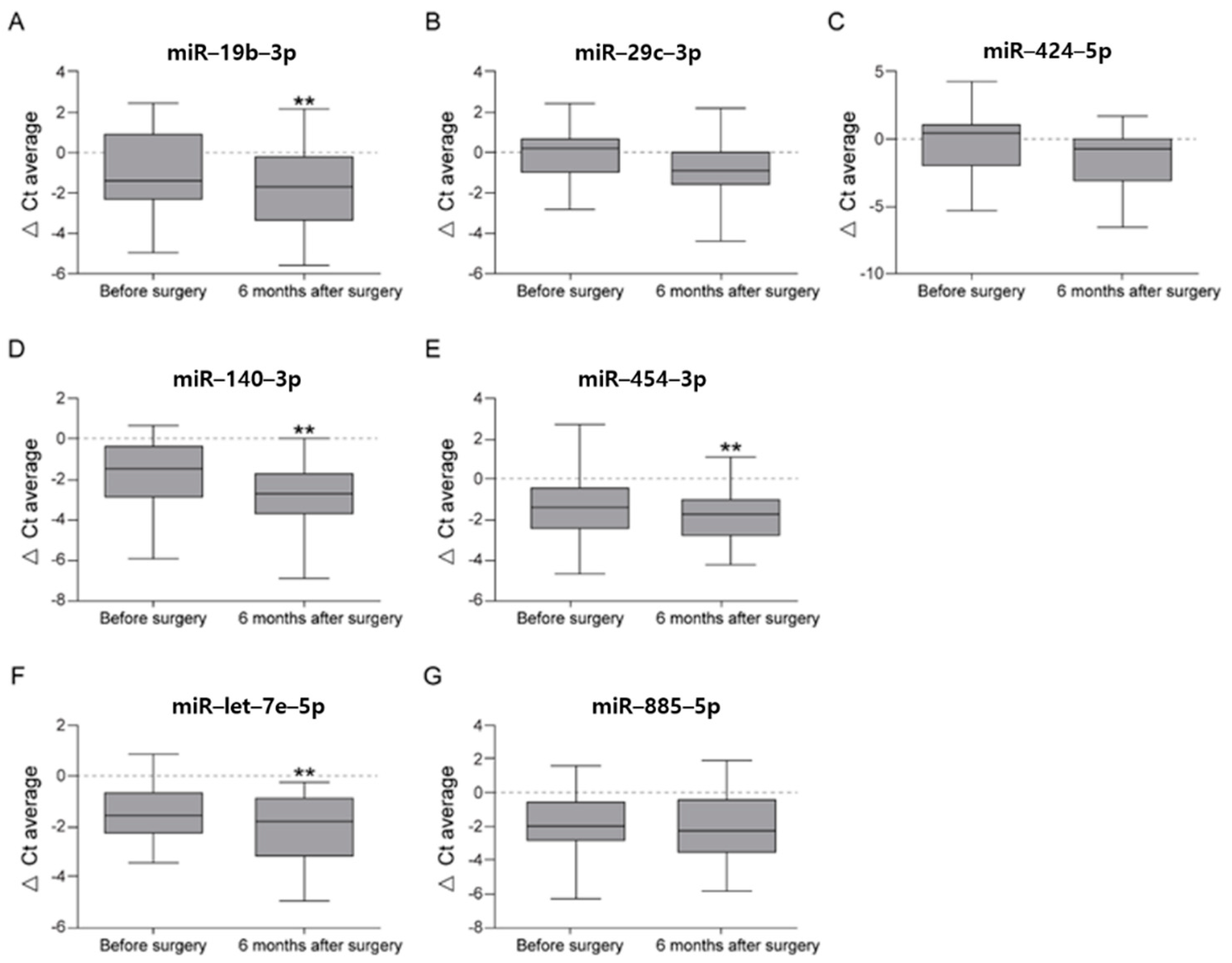
3.2. Validation Stage: Serum Expression Levels of Seven miRNAs at Preoperative and Postoperative Month-6 Time Points in 27 Patients Who Underwent Open Wedge HTO
3.3. Comparison of Clinical and Radiological Outcomes between Preoperative and Postoperative Month-6 Time Points in 27 Patients
3.4. Expression Changes in Seven miRNAs in the Serum 6 and 18 Months after Surgery in 20 Patients Who Were Recruited in the 18-Month Follow-Up Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hunter, D.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Niemeyer, P.; Schmal, H.; Hauschild, O.; Von Heyden, J.; Sudkamp, N.P.; Kostler, W. Open-wedge osteotomy using an internal plate fixator in patients with medial-compartment gonarthritis and varus malalignment: 3-year results with regard to preoperative arthroscopic and radiographic findings. Arthroscopy 2010, 26, 1607–1616. [Google Scholar] [CrossRef]
- Jung, W.H.; Chun, C.W.; Lee, J.H.; Ha, J.H.; Kim, J.H.; Jeong, J.H. Comparative study of medial opening-wedge high tibial osteotomy using 2 different implants. Arthroscopy 2013, 29, 1063–1071. [Google Scholar] [CrossRef]
- Jung, W.H.; Takeuchi, R.; Chun, C.W.; Lee, J.S.; Ha, J.H.; Kim, J.H.; Jeong, J.H. Second-look arthroscopic assessment of cartilage regeneration after medial opening-wedge high tibial osteotomy. Arthroscopy 2014, 30, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Prodromos, C.C.; Amendola, A.; Jakob, R.P. High tibial osteotomy: Indications, techniques, and postoperative management. Instr. Course Lect. 2015, 64, 555–565. [Google Scholar]
- Kanamiya, T.; Naito, M.; Hara, M.; Yoshimura, I. The influences of biomechanical factors on cartilage regeneration after high tibial osteotomy for knees with medial compartment osteoarthritis: Clinical and arthroscopic observation. Arthroscopy 2002, 18, 725–729. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lygrisse, K.; Wang, J. Role of microRNA in osteoarthritis. J. Arthritis 2017, 6, 239. [Google Scholar] [CrossRef]
- Beyer, C.; Zampetaki, A.; Lin, N.Y.; Kleyer, A.; Perricone, C.; Iagnocco, A.; Distler, A.; Langley, S.R.; Gelse, K.; Sesselmann, S.; et al. Signature of circulating microRNAs in osteoarthritis. Ann. Rheum. Dis. 2015, 74, e18. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Bao, J.; Feng, R.; Zhao, Z.; Lu, Q.; Wang, G.; Li, H.; Su, D.; Zhou, J.; Jing, Q.; et al. Circulating microRNAs: A novel potential biomarker for diagnosing acute aortic dissection. Sci. Rep. 2017, 7, 12784. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Zhu, Y.; Tu, G. A bioinformatic analysis of microRNAs role in osteoarthritis. Osteoarthr. Cartil. 2017, 25, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Kung, L.H.W.; Zaki, S.; Ravi, V.; Smith, M.M.; Bell, K.M.; Bateman, J.F.; Little, C.B. Utility of circulating serum miRNAs as biomarkers of early cartilage degeneration in animal models of post-traumatic osteoarthritis and inflammatory arthritis. Osteoarthr. Cartil. 2017, 25, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kirschner, M.B.; van Zandwijk, N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 2011, 80, 193–208. [Google Scholar] [CrossRef]
- Birmingham, T.B.; Moyer, R.; Leitch, K.; Chesworth, B.; Bryant, D.; Willits, K.; Litchfield, R.; Fowler, P.J.; Giffin, J.R. Changes in biomechanical risk factors for knee osteoarthritis and their association with 5-year clinically important improvement after limb realignment surgery. Osteoarthr. Cartil. 2017, 25, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Ruangsomboon, P.; Chareancholvanich, K.; Harnroongroj, T.; Pornrattanamaneewong, C. Survivorship of medial opening wedge high tibial osteotomy in the elderly: Two to ten years of follow up. Int. Orthop. 2017, 41, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Mundermann, A.; Vach, W.; Pagenster, G.; Egloff, C.; Nuesch, C. Assessing in vivo articular cartilage mechanosensitivity as outcome of high tibial osteotomy in patients with medial compartment osteoarthritis: Experimental protocol. Osteoarthr. Cartil. Open 2020, 2, 100043. [Google Scholar] [CrossRef]
- Schuster, P.; Geblein, M.; Schlumberger, M.; Mayer, P.; Mayr, R.; Oremek, D.; Frank, S.; Schulz-Jahrsdorfer, M.; Richter, J. Ten-year results of medial open-wedge high tibial osteotomy and chondral resurfacing in severe medial osteoarthritis and varus malalignment. Am. J. Sports Med. 2018, 46, 1362–1370. [Google Scholar] [CrossRef]
- Kwak, Y.H.; Kwak, D.; Kim, N.Y.; Kim, Y.J.; Lim, J.S.; Yoo, J. Significant changes in synovial fluid microRNAs after high tibial osteotomy in medial compartmental knee osteoarthritis: Identification of potential prognostic biomarkers. PLoS ONE 2020, 15, e0227596. [Google Scholar] [CrossRef] [PubMed]
- Morelanjd, J.R.; Bassett, L.W.; Hanker, G.J. Radiographic analysis of the axial alignment of the lower extremity. J. Bone Jt. Surg. Am. 1987, 69, 745–749. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bignotti, E.; Calza, S.; Tassi, R.A.; Zanotti, L.; Bandiera, E.; Sartori, E.; Odicino, F.E.; Ravaggi, A.; Todeschini, P.; Romani, C. Identification of stably expressed reference small non-coding RNAs for microRNA quantification in high-grade serous ovarian carcinoma tissues. J. Cell. Mol. Med. 2016, 20, 2341–2348. [Google Scholar] [CrossRef]
- Spahn, G.; Hofmann, G.O.; Klinger, H.M. The effects of arthroscopic joint debridement in the knee osteoarthritis: Results of a meta-analysis. Knee Surg. Sports Traumatol. Arthrosc. 2013, 21, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Koh, I.J.; Sohn, S.; Sung, H.S.; In, Y. Degree of preoperative subchondral bone marrow lesion is associated with postoperative outcome after medial opening wedge high tibial osteotomy. Am. J. Sports Med. 2019, 47, 2454–2463. [Google Scholar] [CrossRef]
- Szwedowski, D.; Szczepanek, J.; Paczesny, L.; Pekala, P.; Zabrzynski, J.; Kruczynski, J. Genetics in cartilage lesions: Basic science and therapy approaches. Int. J. Mol. Sci. 2020, 21, 5430. [Google Scholar] [CrossRef] [PubMed]
- Nugent, M. MicroRNAs: Exploring new horizons in osteoarthritis. Osteoarthr. Cartil. 2016, 24, 573–580. [Google Scholar] [CrossRef]
- Lyng, M.B.; Kodahl, A.R.; Binder, H.; Ditzel, H.J. Prospective validation of a blood-based 9-miRNA profile for early detection of breast cancer in a cohort of women examined by clinical mammography. Mol. Oncol. 2016, 10, 1621–1626. [Google Scholar] [CrossRef]
- Kong, R.; Gao, J.; Si, Y.; Zhao, D. Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p expressions correlates with risk and disease severity of knee osteoarthritis. Am. J. Transl. Res. 2017, 9, 2852–2864. [Google Scholar]
- Xu, H.; Liu, X.; Ni, H. Clinical significance of miR-19b-3p in patients with sepsis and its regulatory role in the LPS-induced inflammatory response. Eur. J. Med. Res. 2020, 25, 9. [Google Scholar] [CrossRef]
- Duan, L.; Duan, D.; Wei, W.; Sun, Z.; Xu, H.; Guo, L.; Wu, X. MiR-19b-3p attenuates IL-1β induced extracellular matrix degradation and inflammatory injury in chondrocytes by targeting GRK6. Mol. Cell. Biochem. 2019, 459, 205–214. [Google Scholar] [CrossRef]
- Li, Y.H.; Tavallaee, G.; Tokar, T.; Nakamura, A.; Sundararajan, K.; Weston, A.; Sharma, A.; Mahomed, N.N.; Gandhi, R.; Jurisica, I.; et al. Identification of synovial fluid microRNA signature in knee osteoarthritis. Osteoarthr. Cartil. 2016, 24, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, J.; Millet, M.; Croset, M.; Sornay-Rendu, E.; Borel, O.; Chapurlat, R. Association of circulating microRNAs with prevalent and incident knee osteoarthritis in women: The OFELY study. Arthritis Res. Ther. 2020, 22, 2. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Ma, P.; Wu, N.; Su, X.; Chen, J.; Jiang, C.; Liu, S.; Chen, W.; Ma, B.; Yang, X.; et al. Altered function in cartilage derived mesenchymal stem cell leads to OA-related cartilage erosion. Am. J. Transl. Res. 2016, 8, 433–446. [Google Scholar]
- Yin, C.M.; Suen, W.C.; Lin, S.; Wu, X.M.; Li, G.; Pan, X.H. Dysregulation of both miR-140-39 and miR-140-5p in synovial fluid correlate with osteoarthritis severity. Bone Jt. Res. 2017, 6, 612–618. [Google Scholar] [CrossRef]
- Le, L.T.T.; Swingler, T.E.; Clark, I.M. Review: The role of microRNAs in osteoarthritis and chondrogenesis. Arthritis Rheum. 2013, 65, 1963–1974. [Google Scholar] [CrossRef]
- Lin, L.; Shen, Q.; Zhang, C.; Chen, L.; Yu, C. Assessment of the profiling microRNA expression of differentiated and dedifferentiated human adult articular chondrocytes. J. Orthop. Res. 2011, 29, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Miyaki, S.; Nakasa, T.; Otsuki, S.; Grogan, S.P.; Higashiyama, R.; Inoue, A.; Kato, Y.; Sato, T.; Lotz, M.K.; Asahara, H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009, 60, 2723–2730. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef]
- Ntoumou, E.; Tzetis, M.; Braoudaki, M.; Lambrou, G.; Poulou, M.; Malizos, K.; Stefanou, N.; Anastasopoulou, L.; Tsezou, A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenetics 2017, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.; Rasheed, N.; Al-Shaya, O. Epigallocatechin-3-O-gallate modulates global microRNA expression in interleukin-1β-stimulated human osteoarthritis chondrocytes: Potential role of EGCG on negative co-regulation of microRNA-140-3p and ADAMTS5. Eur. J. Nutr. 2018, 57, 917–928. [Google Scholar] [CrossRef]
- Skrzypa, M.; Szala, D.; Gablo, N.; Czech, J.; Pajak, J.; Kopanska, M.; Trzeciak, M.; Gargasz, K.; Snela, S.; Zawlik, I. miRNA-146a-5p is upregulated in serum and cartilage samples of patients with osteoarthritis. Pol. Prz. Chir. 2019, 91, 1–5. [Google Scholar] [CrossRef]
- Kopańska, M.; Szala, D.; Czech, J.; Gabło, N.; Gargasz, K.; Trzeciak, M.; Zawlik, I.; Snela, S. MiRNA expression in the cartilage of patients with osteoarthritis. J. Orthop. Surg. Res. 2017, 12, 51. [Google Scholar] [CrossRef]
- Shao, N.; Xue, L.; Wang, R.; Luo, K.; Zhi, F.; Lan, Q. miR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol. Cancer Ther. 2019, 18, 459–469. [Google Scholar] [CrossRef]
- Ren, L.; Chen, H.; Song, J.; Chen, X.; Lin, C.; Zhang, X.; Hou, N.; Pan, J.; Zhou, Z.; Wang, L.; et al. MiR-454-3p-mediated Wnt/β-catenin signaling antagonists suppression promotes breast cancer metastasis. Theranostics 2019, 9, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef] [PubMed]
- Templeton, E.M.; Lasse, M.; Pilbrow, A.P.; Cameron, V.A.; Pickering, J.W.; Hertzberg, D.; Frampton, C.M.; Endre, Z.H.; Richards, A.M.; Doughty, R.N.; et al. Circulating microRNA-454-3p is associated with acute kidney injury in patients with acute decompensated heart failure. Genet. Genom. 2019, 140, A13115. [Google Scholar]
- Quintanilha, B.J.; Ferreira, L.R.P.; Ferreira, F.M.; Neto, E.C.; Sampaio, G.R.; Rogero, M.M. Circulating plasma microRNAs dysregulation and metabolic endotoxemia induced by a high-fat high-saturated diet. Clin. Nutr. 2020, 39, 554–562. [Google Scholar] [CrossRef] [PubMed]
| Serial Number | Gender | Age (Year) | Body Mass Index (kg/m2) | Kellgren–Lawrence Grade | Visual Analogue Scale | WOMAC Score | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | Pain | Stiffness | Function | ||||||
| 1 | F | 57 | 28.4 | II | 6 | 54 | 13 | 4 | 37 |
| 2 | F | 55 | 23.9 | II | 5 | 10 | 1 | 0 | 9 |
| 3 | F | 61 | 22.8 | III | 7 | 56 | 8 | 6 | 42 |
| 4 | F | 56 | 27.9 | III | 7 | 42 | 9 | 3 | 30 |
| 5 | F | 65 | 30.2 | III | 3 | 14 | 0 | 0 | 14 |
| 6 | F | 52 | 26.0 | III | 4 | 33 | 9 | 3 | 21 |
| Serial Number (POD 6 mo) | Serial Number (POD 18 mo) | Gender | Age (Year) | Body Mass Index (kg/m2) | Kellgren–Lawrence Grade | Visual Analogue Scale | WOMAC Score | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Pain | Stiffness | Function | |||||||
| 1 | F | 57 | 23.4 | III | 6 | 14 | 3 | 2 | 9 | |
| 2 | 1 | F | 58 | 30.1 | II | 3 | 6 | 2 | 0 | 4 |
| 3 | 2 | F | 50 | 30.5 | III | 3 | 21 | 2 | 2 | 17 |
| 4 | 3 | F | 46 | 33.3 | III | 7 | 22 | 3 | 2 | 17 |
| 5 | F | 54 | 26.5 | II | 7 | 26 | 5 | 2 | 19 | |
| 6 | 4 | F | 51 | 25.5 | II | 9 | 59 | 14 | 3 | 42 |
| 7 | F | 56 | 21.8 | III | 8 | 28 | 5 | 5 | 18 | |
| 8 | F | 57 | 28.4 | II | 6 | 54 | 13 | 4 | 37 | |
| 9 | 5 | F | 55 | 23.9 | II | 5 | 10 | 1 | 0 | 9 |
| 10 | 6 | F | 61 | 22.8 | III | 7 | 56 | 8 | 6 | 42 |
| 11 | 7 | F | 56 | 27.9 | III | 7 | 42 | 9 | 3 | 30 |
| 12 | F | 52 | 26 | III | 4 | 33 | 9 | 3 | 21 | |
| 13 | 8 | M | 64 | 30.9 | III | 7 | 30 | 7 | 2 | 21 |
| 14 | F | 55 | 24 | III | 8 | 48 | 10 | 4 | 34 | |
| 15 | 9 | F | 56 | 26.4 | III | 7 | 52 | 9 | 6 | 37 |
| 16 | 10 | M | 61 | 28.5 | III | 8 | 53 | 12 | 3 | 38 |
| 17 | 11 | F | 59 | 26.3 | III | 5 | 36 | 8 | 3 | 25 |
| 18 | 12 | F | 56 | 27.5 | III | 5 | 42 | 15 | 6 | 21 |
| 19 | 13 | F | 61 | 29.7 | III | 8 | 61 | 13 | 6 | 42 |
| 20 | 14 | F | 57 | 31.2 | III | 6 | 45 | 10 | 4 | 31 |
| 21 | M | 58 | 25.4 | III | 5 | 23 | 7 | 3 | 13 | |
| 22 | 15 | F | 56 | 21.8 | III | 3 | 29 | 5 | 2 | 22 |
| 23 | 16 | F | 56 | 25.4 | III | 2 | 26 | 5 | 2 | 19 |
| 24 | 17 | M | 59 | 24.8 | III | 8 | 37 | 7 | 4 | 26 |
| 25 | 18 | F | 60 | 25.2 | III | 7 | 37 | 2 | 4 | 41 |
| 26 | 19 | M | 64 | 25.7 | III | 7 | 42 | 5 | 0 | 37 |
| 27 | 20 | M | 56 | 25.4 | III | 4 | 25 | 4 | 3 | 18 |
| miRNA Symbol | Assay ID | Catalogue No. | Sequencing |
|---|---|---|---|
| SNORD61 | Hs_SNORD61_11 | MS00033705 | |
| miR-19b-3p | Hs_miR-19b_2 | MS00031584 | 5′UGUGCAAAUCCAUGCAAAACUGA |
| miR-29c-3p | Hs_miR-29c_1 | MS00003269 | 5′UAGCACCAUUUGAAAUCGGUUA |
| miR-424-5p | Hs_miR-424_1 | MS00004186 | 5′CAGCAGCAAUUCAUGUUUUGAA |
| miR-140-3p | Hs_miR-140-3p_1 | MS00008673 | 5′UACCACAGGGUAGAACCACGG |
| miR-454-3p | Hs_miR-454_1 | MS00007861 | 5′UAGUGCAAUAUUGCUUAUAGGGU |
| miR-let-7e-5p | Hs_let-7e_3 | MS00031227 | 5′UGAGGUAGGAGGUUGUAUAGUU |
| miR-885-5p | Hs_miR-885-5p_1 | MS00010668 | 5′UCCAUUACACUACCCUGCCUCU |
| Procedure | Chemical/Assay Kit |
|---|---|
| RNA preparation | miRNeasy Serum/Plasma kit (Qiagen) |
| miRNeasy Serum/Plasma Spike-In Control (Qiagen) | |
| QIAzol Lysis Reagent (Qiagen) | |
| Absolute Ethanol (Merck) | |
| Chloroform (without added isoamyl alcohol) (Sigma) | |
| PCR array | miScript miRNA PCR array (MIHS-105Z) (Qiagen) |
| Reverse Transcription | miScript II RT kit (Qiagen) |
| Real-Time PCR | miScript SYBR Green PCR kit (Qiagen) Specific primer assays (Table 3) |
| miRNA | Preop. | Postop. Month 6 | Fold Regulation | p-Value * | ||
|---|---|---|---|---|---|---|
| ΔCt | 2−ΔCt | ΔCt | 2−ΔCt | |||
| hsa-miR-19b-3p | −0.66 ± 0.87 | 1.57 | −2.47 ± 1.38 | 5.53 | 3.51 | 0.011 |
| hsa-miR-29c-3p | −0.32 ± 0.79 | 1.25 | −2.10 ± 1.26 | 4.28 | 3.43 | 0.015 |
| hsa-miR-424-5p | −0.48 ± 1.17 | 1.39 | −2.23 ± 0.80 | 4.69 | 3.37 | 0.021 |
| miRNA | Profiling Stage Using Microarray | p-Values * | Validation Stage Using Real-Time PCR | p-Values * |
|---|---|---|---|---|
| Fold Change | Fold Change | |||
| has-miR-19b-3p | 3.51 | 0.011 | 3.66 | 0.003 ** |
| has-miR-29c-3p | 3.43 | 0.015 | 3.68 | 0.01 |
| has-miR-424-5p | 3.37 | 0.021 | 5.49 | 0.063 |
| has-miR-140-3p | - | - | 4.12 | 0.005 ** |
| has-miR-454-3p | - | - | 2.18 | 0.004 ** |
| has-miR-let-7e-5p | - | - | 2.38 | 0.004 ** |
| has-miR-885-5p | - | - | 2.06 | 0.096 |
| Variables | Preoperative | Postoperative Month-6 | p-Value * |
|---|---|---|---|
| Mechanical axis (−: varus, +: valgus) | −5.0 ± 2.2 | +2.5 ± 1.5 | <0.001 |
| Weight-bearing line ratio (%) | 18.6 ± 6.2 | 59.5 ± 8.2 | <0.001 |
| Visual analogue scale | 6.0 ± 1.9 | 2.6 ± 1.1 | <0.001 |
| WOMAC total | 36.4 ± 16.0 | 24.2 ± 11.1 | 0.002 |
| Pain | 7.8 ± 4.0 | 4.3 ± 2.5 | 0.001 |
| Stiffness | 3.2 ± 1.7 | 2.2 ± 1.3 | 0.022 |
| Function | 25.4 ± 11.4 | 17.8 ± 8.5 | 0.006 |
| miRNA | 6-Month | 18-Month | p-Value * |
|---|---|---|---|
| Fold Change | Fold Change | ||
| has-miR-19b-3p | 2.99 | 5.91 | 0.012 |
| has-miR-29c-3p | 2.3 | 4.44 | 0.016 |
| has-miR-424-5p | 5.13 | 8.6 | 0.003 * |
| has-miR-140-3p | 3.09 | 8.48 | 0.003 * |
| has-miR-454-3p | 2.6 | 3.57 | 0.027 |
| has-miR-let-7e-5p | 1.25 | 2.38 | 0.071 |
| has-miR-885-5p | 1.31 | 2.93 | 0.153 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwak, Y.H.; Kwak, D.-K.; Moon, H.-S.; Kim, N.Y.; Yee, J.-S.; Yoo, J.-H. Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers. Diagnostics 2021, 11, 258. https://doi.org/10.3390/diagnostics11020258
Kwak YH, Kwak D-K, Moon H-S, Kim NY, Yee J-S, Yoo J-H. Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers. Diagnostics. 2021; 11(2):258. https://doi.org/10.3390/diagnostics11020258
Chicago/Turabian StyleKwak, Yoon Hae, Dae-Kyung Kwak, Hyun-Soo Moon, Nan Young Kim, Jae-Sung Yee, and Je-Hyun Yoo. 2021. "Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers" Diagnostics 11, no. 2: 258. https://doi.org/10.3390/diagnostics11020258
APA StyleKwak, Y. H., Kwak, D.-K., Moon, H.-S., Kim, N. Y., Yee, J.-S., & Yoo, J.-H. (2021). Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers. Diagnostics, 11(2), 258. https://doi.org/10.3390/diagnostics11020258






