Quantitative Evaluation of CFTR Pre-mRNA Splicing Dependent on the (TG)mTn Poly-Variant Tract
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lucarelli, M.; Porcaro, L.; Biffignandi, A.; Costantino, L.; Giannone, V.; Alberti, L.; Bruno, S.M.; Corbetta, C.; Torresani, E.; Colombo, C.; et al. A New Targeted CFTR Mutation Panel Based on Next-Generation Sequencing Technology. J. Mol. Diagn. 2017, 19, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Ferraguti, G.; Pierandrei, S.; Bruno, S.M.; Ceci, F.; Strom, R.; Lucarelli, M. A template for mutational data analysis of the CFTR gene. Clin. Chem. Lab. Med. 2011, 49, 1447–1451. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ivanov, M.; Matsvay, A.; Glazova, O.; Krasovskiy, S.; Usacheva, M.; Amelina, E.; Chernyak, A.; Ivanov, M.; Musienko, S.; Prodanov, T.; et al. Targeted sequencing reveals complex, phenotype-correlated genotypes in cystic fibrosis. BMC Med Genom. 2018, 11, 13. [Google Scholar] [CrossRef]
- Bergougnoux, A.; D’Argenio, V.; Sollfrank, S.; Verneau, F.; Telese, A.; Postiglione, I.; Lackner, K.J.; Claustres, M.; Castaldo, G.; Rossmann, H.; et al. Multicenter validation study for the certification of a CFTR gene scanning method using next generation sequencing technology. Clin. Chem. Lab. Med. 2018, 56, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Bergougnoux, A.; Lopez, M.; Girodon, E. The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis. Int. J. Neonatal Screen. 2020, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Bergougnoux, A.; Taulan-Cadars, M.; Claustres, M.; Raynal, C. Current and future molecular approaches in the diagnosis of cystic fibrosis. Expert Rev. Respir. Med. 2018, 12, 415–426. [Google Scholar] [CrossRef]
- Straniero, L.; Soldà, G.; Costantino, L.; Seia, M.; Melotti, P.; Colombo, C.; Asselta, R.; Duga, S. Whole-gene CFTR sequencing combined with digital RT-PCR improves genetic diagnosis of cystic fibrosis. J. Hum. Genet. 2016, 61, 977–984. [Google Scholar] [CrossRef]
- Lucarelli, M.; Bruno, S.M.; Pierandrei, S.; Ferraguti, G.; Testino, G.; Truglio, G.; Strom, R.; Quattrucci, S. The Impact on Genetic Testing of Mutational Patterns of CFTR Gene in Different Clinical Macrocategories of Cystic Fibrosis. J. Mol. Diagn. 2016, 18, 554–565. [Google Scholar] [CrossRef][Green Version]
- Terlizzi, V.; Piccialli, G.; Salvatore, D.; Lucarelli, M.; Raia, V.; Angioni, A.; Carnovale, V.; Cirilli, N.; Casciaro, R.; Colombo, C.; et al. Genotype–phenotype correlation and functional studies in patients with cystic fibrosis bearing CFTR complex alleles. J. Med. Genet. 2017, 54, 224–235. [Google Scholar] [CrossRef]
- Tsui, L.-C.; Dorfman, R. The Cystic Fibrosis Gene: A Molecular Genetic Perspective. Cold Spring Harb. Perspect. Med. 2013, 3, a009472. [Google Scholar] [CrossRef]
- Cutting, G.R. Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 2015, 16, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Terlizzi, V.; Lucarelli, M.; Salvatore, D.; Angioni, A.; Bisogno, A.; Braggion, C.; Buzzetti, R.; Carnovale, V.; Casciaro, R.; Castaldo, G.; et al. Clinical expression of cystic fibrosis in a large cohort of Italian siblings. BMC Pulm. Med. 2018, 18, 196. [Google Scholar] [CrossRef] [PubMed]
- Terlizzi, V.; Carnovale, V.; Piccialli, G.; Castellani, C.; Cirilli, N.; Colombo, C.; Corti, F.; Cresta, F.; D’Adda, A.; Lucarelli, M.; et al. Clinical expression of patients with the D1152H CFTR mutation. J. Cyst. Fibros. 2015, 14, 447–452. [Google Scholar] [CrossRef]
- Narzi, L.; Ferraguti, G.; Stamato, A.; Narzi, F.; Valentini, S.; Lelli, A.; Delaroche, I.; Lucarelli, M.; Strom, R.; Quattrucci, S. Does cystic fibrosis neonatal screening detect atypical CF forms? Extended genetic characterization and 4-year clinical follow-up. Clin. Genet. 2007, 72, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Bareil, C.; Guittard, C.; Altieri, J.-P.; Templin, C.; Claustres, M.; Georges, M.D. Comprehensive and Rapid Genotyping of Mutations and Haplotypes in Congenital Bilateral Absence of the Vas Deferens and Other Cystic Fibrosis Transmembrane Conductance Regulator-Related Disorders. J. Mol. Diagn. 2007, 9, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Bobadilla, J.L.; Macek, M.; Fine, J.P.; Farrell, P.M. Cystic fibrosis: A worldwide analysis ofCFTR mutations?correlation with incidence data and application to screening. Hum. Mutat. 2002, 19, 575–606. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, K.A.; Taber, K.A.J.; Grauman, P.V.; Spurka, L.; Lim-Harashima, J.; Svenson, A.; Goldberg, J.D.; Muzzey, D. Sequencing as a first-line methodology for cystic fibrosis carrier screening. Genet. Med. 2019, 21, 2569–2576. [Google Scholar] [CrossRef]
- Bonadia, L.C.; Marson, F.A.L.; Ribeiro, J.D.; Paschoal, I.A.; Pereira, M.C.; Ribeiro, A.F.; Battagin, A.S. CFTR genotype and clinical outcomes of adult patients carried as cystic fibrosis disease. Gene 2014, 540, 183–190. [Google Scholar] [CrossRef]
- Bear, C.E. A Therapy for Most with Cystic Fibrosis. Cell 2020, 180, 211. [Google Scholar] [CrossRef]
- Amaral, M.D. Novel personalized therapies for cystic fibrosis: Treating the basic defect in all patients. J. Intern. Med. 2015, 277, 155–166. [Google Scholar] [CrossRef]
- Rossi, T.; Grandoni, F.; Mazzilli, F.; Quattrucci, S.; Antonelli, M.; Strom, R.; Lucarelli, M. High frequency of (TG)mTn variant tracts in the cystic fibrosis transmembrane conductance regulator gene in men with high semen viscosity. Fertil. Steril. 2004, 82, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, M.; Bruno, S.M.; Pierandrei, S.; Ferraguti, G.; Stamato, A.; Narzi, F.; Amato, A.; Cimino, G.; Bertasi, S.; Quattrucci, S.; et al. A Genotypic-Oriented View of CFTR Genetics Highlights Specific Mutational Patterns Underlying Clinical Macrocategories of Cystic Fibrosis. Mol. Med. 2015, 21, 257–275. [Google Scholar] [CrossRef]
- Claustres, M. Molecular pathology of the CFTR locus in male infertility. Reprod. Biomed. Online 2005, 10, 14–41. [Google Scholar] [CrossRef]
- Stuhrmann, M.; Dörk, T. CFTR gene mutations and male infertility. Andrologia 2000, 32, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Pierandrei, S.; Blacona, G.; Fabrizzi, B.; Cimino, G.; Cirilli, N.; Caporelli, N.; Angeloni, A.; Cipolli, M.; Lucarelli, M. Two novel and correlated CF-causing insertions in the (TG)mTn tract of the CFTR gene. PLoS ONE 2019, 14, e0222838. [Google Scholar] [CrossRef]
- Giordano, S.; Amato, F.; Elce, A.; Monti, M.; Iannone, C.; Pucci, P.; Seia, M.; Angioni, A.; Zarrilli, F.; Castaldo, G.; et al. Molecular and Functional Analysis of the Large 5′ Promoter Region of CFTR Gene Revealed Pathogenic Mutations in CF and CFTR-Related Disorders. J. Mol. Diagn. 2013, 15, 331–340. [Google Scholar] [CrossRef]
- Tomaiuolo, A.C.; Alghisi, F.; Petrocchi, S.; Surace, C.; Roberti, M.C.; Bella, S.; Lucidi, V.; Angioni, A. Clinical hallmarks and genetic polymorphisms in the CFTR gene contribute to the disclosure of the A1006E mutation. Clin. Investig. Med. 2010, 33, E234–E239. [Google Scholar] [CrossRef]
- Pagin, A.; Devos, A.; Figeac, M.; Truant, M.; Willoquaux, C.; Broly, F.; Lalau, G. Applicability and Efficiency of NGS in Routine Diagnosis: In-Depth Performance Analysis of a Complete Workflow for CFTR Mutation Analysis. PLoS ONE 2016, 11, e0149426. [Google Scholar] [CrossRef]
- Kiesewetter, S.; Macek, M.; Davis, C.; Curristin, S.M.; Chu, C.-S.; Graham, C.; Shrimpton, A.E.; Cashman, S.M.; Tsui, L.-C.; Mickle, J.; et al. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat. Genet. 1993, 5, 274–278. [Google Scholar] [CrossRef]
- Massie, R.; Poplawski, N.; Wilcken, B.; Goldblatt, J.; Byrnes, C.; Robertson, C. Intron-8 polythymidine sequence in Australasian individuals with CF mutations R117H and R117C. Eur. Respir. J. 2001, 17, 1195–1200. [Google Scholar] [CrossRef]
- Feldmann, D.; Couderc, R.; Audrezet, M.-P.; Ferec, C.; Bienvenu, T.; Desgeorges, M.; Claustres, M.; Blayau, M.; Bozon, D.; Malinge, M.-C.; et al. CFTR genotypes in patients with normal or borderline sweat chloride levels. Hum. Mutat. 2003, 22, 340. [Google Scholar] [CrossRef] [PubMed]
- Cuppens, H.; Lin, W.; Jaspers, M.; Costes, B.; Teng, H.; Vankeerberghen, A.; Jorissen, M.; Droogmans, G.; Reynaert, I.; Goossens, M.; et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J. Clin. Investig. 1998, 101, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Niksic, M.; Romano, M.; Buratti, E.; Pagani, F.; Baralle, F.E. Functional Analysis of cis-Acting Elements Regulating the Alternative Splicing of Human CFTR Exon 9. Hum. Mol. Genet. 1999, 8, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, M.; Grandoni, F.; Rossi, T.; Mazzilli, F.; Antonelli, M.; Strom, R. Simultaneous Cycle Sequencing Assessment of (TG)m and Tn Tract Length in CFTR Gene. Biotechniques 2002, 32, 540–547. [Google Scholar] [CrossRef]
- Lucarelli, M.; Narzi, L.; Piergentili, R.; Ferraguti, G.; Grandoni, F.; Quattrucci, S.; Strom, R. A 96-well formatted method for exon and exon/intron boundary full sequencing of the CFTR gene. Anal. Biochem. 2006, 353, 226–235. [Google Scholar] [CrossRef]
- Chillón, M.; Casals, T.; Mercier, B.; Bassas, L.; Lissens, W.; Silber, S.; Romey, M.-C.; Ruiz-Romero, J.; Verlingue, C.; Claustres, M.; et al. Mutations in the Cystic Fibrosis Gene in Patients with Congenital Absence of the Vas Deferens. N. Engl. J. Med. 1995, 332, 1475–1480. [Google Scholar] [CrossRef]
- Zhang, L.; Button, B.; Gabriel, S.E.; Burkett, S.; Yan, Y.; Skiadopoulos, M.H.; Dang, Y.L.; Vogel, L.N.; McKay, T.; Mengos, A.; et al. CFTR Delivery to 25% of Surface Epithelial Cells Restores Normal Rates of Mucus Transport to Human Cystic Fibrosis Airway Epithelium. PLoS Biol. 2009, 7, e1000155. [Google Scholar] [CrossRef]
- Buratti, E.; Brindisi, A.; Pagani, F.; Fe, B. Nuclear Factor TDP-43 Binds to the Polymorphic TG Repeats in CFTR Intron 8 and Causes Skipping of Exon 9: A Functional Link with Disease Penetrance. Am. J. Hum. Genet. 2004, 74, 1322–1325. [Google Scholar] [CrossRef]
- Pagani, F.; Buratti, E.; Stuani, C.; Romano, M.; Zuccato, E.; Niksic, M.; Giglio, L.; Faraguna, D.; Baralle, F.E. Splicing Factors Induce Cystic Fibrosis Transmembrane Regulator Exon 9 Skipping through a Nonevolutionary Conserved Intronic Element. J. Biol. Chem. 2000, 275, 21041–21047. [Google Scholar] [CrossRef]
- Pagani, F.; Baralle, F.E. Genomic variants in exons and introns: Identifying the splicing spoilers. Nat. Rev. Genet. 2004, 5, 389–396. [Google Scholar] [CrossRef]
- Amaral, M.D.; Clarke, L.A.; Ramalho, A.; Beck, S.; Broackes-Carter, F.; Rowntree, R.; Mouchel, N.; Williams, S.H.; Harris, A.; Tzetis, M.; et al. Quantitative methods for the analysis of CFTR transcripts/splicing variants. J. Cyst. Fibros. 2004, 3 (Suppl. 2), 17–23. [Google Scholar] [CrossRef] [PubMed]
- Andrieux, J.; Audrézet, M.P.; Frachon, I.; Leroyer, C.; Roge, C.; Scotet, V.; Férec, C. Quantification of CFTR splice variants in adults with disseminated bronchiectasis, using the TaqMan fluorogenic detection system. Clin. Genet. 2002, 62, 60–67. [Google Scholar] [CrossRef] [PubMed]

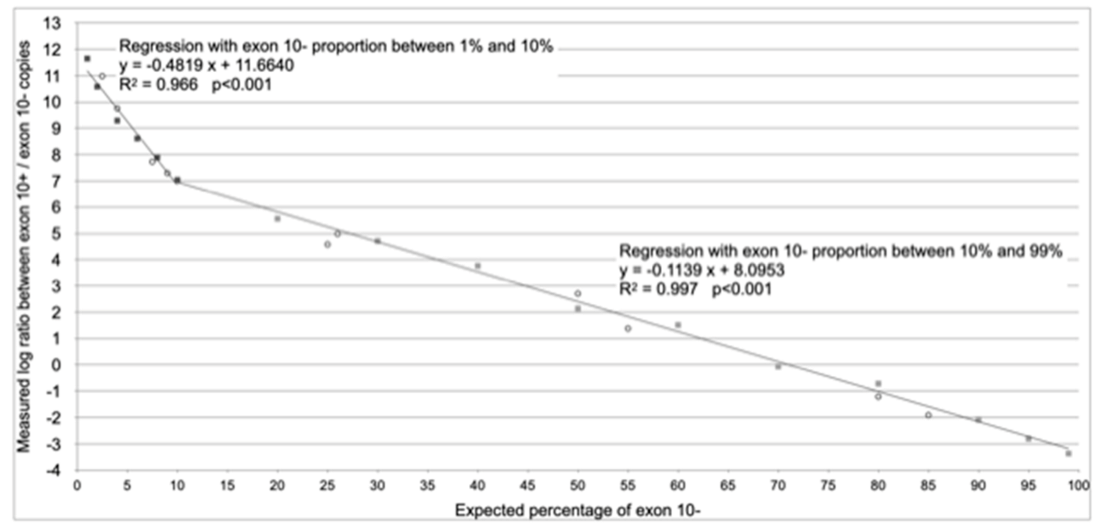
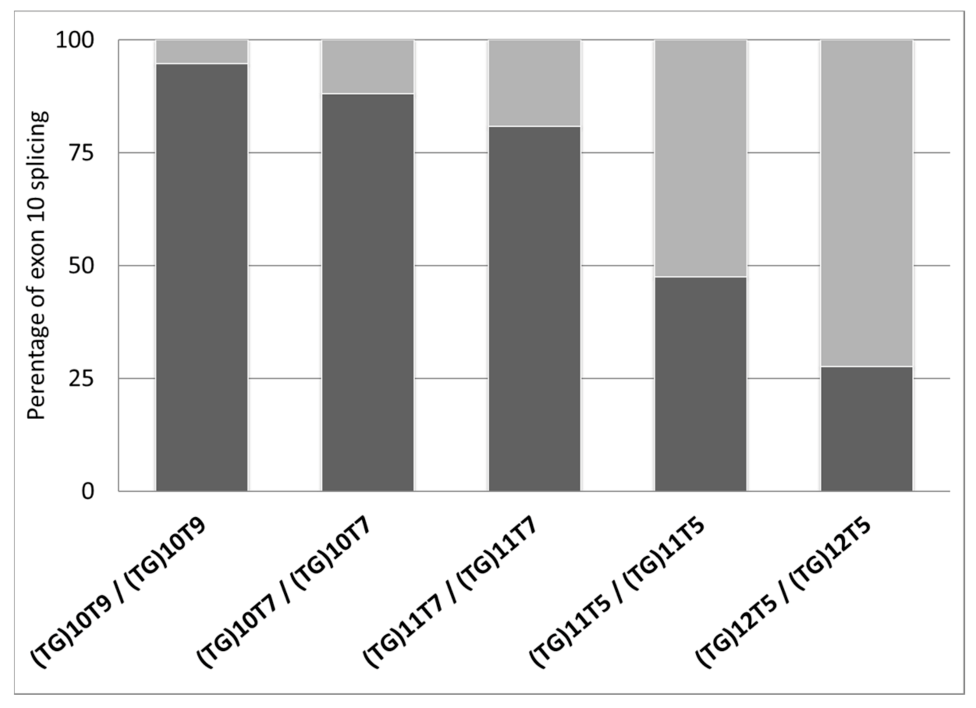
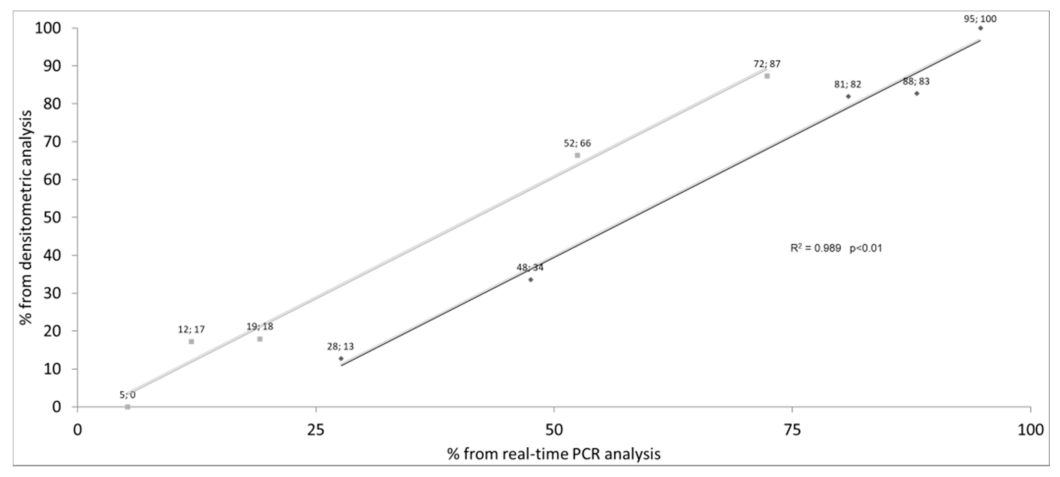
| ID | Diagnosis | Genotype (Legacy Name) | (TG)mTn (Legacy Name) | Exon 10 Splicing Percentages | |||
|---|---|---|---|---|---|---|---|
| Individual Percentages | Average Percentages ± ds | ||||||
| Exon 10+ | Exon 10− | Exon 10+ | Exon 10− | ||||
| 1 | CF-PS | F508del/1585-9412A>G * | (TG)10T9/(TG)10T9 | 97.8 | 2.2 | 94.7 ± 2.7 | 5.3 ± 2.7 |
| 2 | CF-PI | F508del/F508del | (TG)10T9/(TG)10T9 | 93.5 | 6.5 | ||
| 3 | CF-PI | [F508del;I1027T]/F508del | (TG)10T9/(TG)10T9 | 92.9 | 7.1 | ||
| 4 | Uncertain | G576A/G576A | (TG)10T7/(TG)10T7 | 91.3 | 7.7 | 88.0 ± 4.6 | 12.0 ± 4.6 |
| 5 | Healthy (carrier) | CFTRdup19/+ | (TG)10T7/(TG)10T7 | 84.7 | 15.3 | ||
| 6 | CF-PI | R553X/CFTRdele2 | (TG)11T7/(TG)11T7 | 83.1 | 16.9 | 80.9 ± 2.2 | 19.1 ± 2.2 |
| 7 | CFTR-RD | unknown/unknown | (TG)11T7/(TG)11T7 | 81.4 | 18.6 | ||
| 8 | Healthy (carrier) | R553X/+ | (TG)11T7/(TG)11T7 | 79.6 | 20.4 | ||
| 9 | Healthy (carrier) | G85E/+ | (TG)11T7/(TG)11T7 | 77.7 | 22.3 | ||
| 10 | Healthy (carrier) | G85E/+ | (TG)11T7/(TG)11T7 | 82.4 | 17.6 | ||
| 11 | Healthy (gen pop) | +/+ | (TG)11T5/(TG)11T5 | 47.5 | 52.5 | 47.5 ± 0.0 | 52.5 ± 0.0 |
| 12 | CFTR-RD | 359insT/+ | (TG)12T5/(TG)12T5 | 36.2 | 63.8 | 27.6 ± 12.1 | 72.4 ± 12.1 |
| 13 | CFTR-RD | +/+ | (TG)12T5/(TG)12T5 | 19.1 | 80.9 | ||
| Range of Applicability | From 1% to 10% of Splicing without Exon 10 (Exon 10− Assay) | From 10% to 99% of Splicing without Exon 10 (Exon 10− Assay) |
|---|---|---|
| Regression equation (according to Figure 2) | y = −0.4819 x + 11.6640 | y = −0.1139 x + 8.0953 |
| Reverse regression equation (x = % of splicing without exon 10) | x = (y − 11.6640)/−0.4819 | x = (y − 8.0953)/−0.1139 |
| Formula with generic efficiency | % splicing without exon 10 = | % splicing without exon 10 = |
| Formula with experimental values of effciencies | % splicing without exon 10 = | % splicing without exon 10 = |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sterrantino, M.; Fuso, A.; Pierandrei, S.; Bruno, S.M.; Testino, G.; Cimino, G.; Angeloni, A.; Lucarelli, M. Quantitative Evaluation of CFTR Pre-mRNA Splicing Dependent on the (TG)mTn Poly-Variant Tract. Diagnostics 2021, 11, 168. https://doi.org/10.3390/diagnostics11020168
Sterrantino M, Fuso A, Pierandrei S, Bruno SM, Testino G, Cimino G, Angeloni A, Lucarelli M. Quantitative Evaluation of CFTR Pre-mRNA Splicing Dependent on the (TG)mTn Poly-Variant Tract. Diagnostics. 2021; 11(2):168. https://doi.org/10.3390/diagnostics11020168
Chicago/Turabian StyleSterrantino, Manuela, Andrea Fuso, Silvia Pierandrei, Sabina Maria Bruno, Giancarlo Testino, Giuseppe Cimino, Antonio Angeloni, and Marco Lucarelli. 2021. "Quantitative Evaluation of CFTR Pre-mRNA Splicing Dependent on the (TG)mTn Poly-Variant Tract" Diagnostics 11, no. 2: 168. https://doi.org/10.3390/diagnostics11020168
APA StyleSterrantino, M., Fuso, A., Pierandrei, S., Bruno, S. M., Testino, G., Cimino, G., Angeloni, A., & Lucarelli, M. (2021). Quantitative Evaluation of CFTR Pre-mRNA Splicing Dependent on the (TG)mTn Poly-Variant Tract. Diagnostics, 11(2), 168. https://doi.org/10.3390/diagnostics11020168








