Quantification of Sodium Relaxation Times and Concentrations as Surrogates of Proteoglycan Content of Patellar CARTILAGE at 3T MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. MRI
2.2.1. Characterization of the 23Na Coil
2.2.2. MRI Sequence Parameters
2.3. Image Post-Processing
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MacKay, J.W.; Low, S.B.L.; Smith, T.O.; Toms, A.P.; McCaskie, A.W.; Gilbert, F.J. Systematic Review and Meta-Analysis of the Reliability and Discriminative Validity of Cartilage Compositional MRI in Knee Osteoarthritis. Osteoarthr. Cartil. 2018, 26, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Sewerin, P.; Müller-Lutz, A.; Odendahl, S.; Eichner, M.; Schneider, M.; Ostendorf, B.; Schleich, C. Prevention of the Progressive Biochemical Cartilage Destruction under Methotrexate Therapy in Early Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2018, 37, 179–185. [Google Scholar] [CrossRef]
- Abrar, D.B.; Schleich, C.; Frenken, M.; Vordenbäumen, S.; Richter, J.; Schneider, M.; Ostendorf, B.; Nebelung, S.; Sewerin, P. DGEMRIC in the Assessment of Pre-Morphological Cartilage Degeneration in Rheumatic Disease: Rheumatoid Arthritis vs. Psoriatic Arthritis. Diagnostics 2021, 11, 147. [Google Scholar] [CrossRef]
- Abrar, D.B.; Schleich, C.; Nebelung, S.; Frenken, M.; Ullrich, T.; Radke, K.L.; Antoch, G.; Vordenbäumen, S.; Brinks, R.; Schneider, M.; et al. Proteoglycan Loss in the Articular Cartilage Is Associated with Severity of Joint Inflammation in Psoriatic Arthritis—a Compositional Magnetic Resonance Imaging Study. Arthritis Res. Ther. 2020, 22, 124. [Google Scholar] [CrossRef] [PubMed]
- Matzat, S.J.; van Tiel, J.; Gold, G.E.; Oei, E.H.G. Quantitative MRI Techniques of Cartilage Composition. Quant. Imaging Med. Surg. 2013, 3, 162–174. [Google Scholar] [CrossRef]
- Bittersohl, B.; Kircher, J.; Miese, F.R.; Dekkers, C.; Habermeyer, P.; Fröbel, J.; Antoch, G.; Krauspe, R.; Zilkens, C. T2* Mapping and Delayed Gadolinium-Enhanced Magnetic Resonance Imaging in Cartilage (DGEMRIC) of Humeral Articular Cartilage-a Histologically Controlled Study. J. Shoulder Elb. Surg. 2015, 24, 1644–1652. [Google Scholar] [CrossRef]
- Besselink, N.J.; Vincken, K.L.; Bartels, L.W.; van Heerwaarden, R.J.; Concepcion, A.N.; Marijnissen, A.C.A.; Spruijt, S.; Custers, R.J.H.; van der Woude, J.-T.A.D.; Wiegant, K.; et al. Cartilage Quality (DGEMRIC Index) Following Knee Joint Distraction or High Tibial Osteotomy. Cartilage 2020, 11, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Schleich, C.; Bittersohl, B.; Miese, F.; Schmitt, B.; Müller-Lutz, A.; Sondern, M.; Antoch, G.; Krauspe, R.; Zilkens, C. Glycosaminoglycan Chemical Exchange Saturation Transfer at 3T MRI in Asymptomatic Knee Joints. Acta Radiol. 2016, 57, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Abrar, D.B.; Schleich, C.; Radke, K.L.; Frenken, M.; Stabinska, J.; Ljimani, A.; Wittsack, H.-J.; Antoch, G.; Bittersohl, B.; Hesper, T.; et al. Detection of Early Cartilage Degeneration in the Tibiotalar Joint Using 3 T GagCEST Imaging: A Feasibility Study. Magn. Reson. Mater. Phys. Biol. Med. 2021, 34, 249–260. [Google Scholar] [CrossRef]
- Krishnamoorthy, G.; Nanga, R.P.R.; Bagga, P.; Hariharan, H.; Reddy, R. High Quality Three-dimensional GagCEST Imaging of in Vivo Human Knee Cartilage at 7 Tesla. Magn. Reson. Med. 2017, 77, 1866–1873. [Google Scholar] [CrossRef]
- Madelin, G.; Xia, D.; Brown, R.; Babb, J.; Chang, G.; Krasnokutsky, S.; Regatte, R.R. Longitudinal Study of Sodium MRI of Articular Cartilage in Patients with Knee Osteoarthritis: Initial Experience with 16-Month Follow-Up. Eur. Radiol. 2018, 28, 133–142. [Google Scholar] [CrossRef]
- Zbýň, Š.; Schreiner, M.; Juras, V.; Mlynarik, V.; Szomolanyi, P.; Laurent, D.; Scotti, C.; Haber, H.; Deligianni, X.; Bieri, O.; et al. Assessment of Low-Grade Focal Cartilage Lesions in the Knee With Sodium MRI at 7 T. Investig. Radiol. 2020, 55, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Zbýň, Š.; Brix, M.O.; Juras, V.; Domayer, S.E.; Walzer, S.M.; Mlynarik, V.; Apprich, S.; Buckenmaier, K.; Windhager, R.; Trattnig, S. Sodium Magnetic Resonance Imaging of Ankle Joint in Cadaver Specimens, Volunteers, and Patients After Different Cartilage Repair Techniques at 7 T. Investig. Radiol. 2015, 50, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, A.; Mellon, E.; Niyogi, S.; Witschey, W.; Kneeland, J.B.; Reddy, R. Sodium AndT1ρ MRI for Molecular and Diagnostic Imaging of Articular Cartilage. NMR Biomed. 2006, 19, 781–821. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Madelin, G.; Sherman, O.H.; Strauss, E.J.; Xia, D.; Recht, M.P.; Jerschow, A.; Regatte, R.R. Improved Assessment of Cartilage Repair Tissue Using Fluid-Suppressed 23Na Inversion Recovery MRI at 7 Tesla: Preliminary Results. Eur. Radiol. 2012, 22, 1341–1349. [Google Scholar] [CrossRef]
- Insko, E.K.; Kaufman, J.H.; Leigh, J.S.; Reddy, R. Sodium NMR Evaluation of Articular Cartilage Degradation. Magn. Reson. Med. 1999, 41, 30–34. [Google Scholar] [CrossRef]
- Ladd, M.E.; Bachert, P.; Meyerspeer, M.; Moser, E.; Nagel, A.M.; Norris, D.G.; Schmitter, S.; Speck, O.; Straub, S.; Zaiss, M. Pros and Cons of Ultra-High-Field MRI/MRS for Human Application. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 109, 1–50. [Google Scholar] [CrossRef]
- Madelin, G.; Lee, J.-S.; Regatte, R.R.; Jerschow, A. Sodium MRI: Methods and Applications. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 79, 14–47. [Google Scholar] [CrossRef]
- Madelin, G.; Regatte, R.R. Biomedical Applications of Sodium MRI in Vivo. J. Magn. Reson. Imaging 2013, 38, 511–529. [Google Scholar] [CrossRef]
- Feldman, R.E.; Stobbe, R.; Watts, A.; Beaulieu, C. Sodium Imaging of the Human Knee Using Soft Inversion Recovery Fluid Attenuation. J. Magn. Reson. 2013, 234, 197–206. [Google Scholar] [CrossRef]
- Staroswiecki, E.; Bangerter, N.K.; Gurney, P.T.; Grafendorfer, T.; Gold, G.E.; Hargreaves, B.A. In Vivo Sodium Imaging of Human Patellar Cartilage with a 3D Cones Sequence at 3 T and 7 T. J. Magn. Reson. Imaging 2010, 32, 446–451. [Google Scholar] [CrossRef]
- Rong, P.; Regatte, R.R.; Jerschow, A. Clean Demarcation of Cartilage Tissue 23Na by Inversion Recovery. J. Magn. Reson. 2008, 193, 207–209. [Google Scholar] [CrossRef]
- Lee, J.-S.; Xia, D.; Madelin, G.; Regatte, R.R. Sodium Inversion Recovery MRI on the Knee Joint at 7 T with an Optimal Control Pulse. J. Magn. Reson. 2016, 262, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Madelin, G.; Babb, J.; Xia, D.; Chang, G.; Krasnokutsky, S.; Abramson, S.B.; Jerschow, A.; Regatte, R.R. Articular Cartilage: Evaluation with Fluid-Suppressed 7.0-T Sodium MR Imaging in Subjects with and Subjects without Osteoarthritis. Radiology 2013, 268, 481–491. [Google Scholar] [CrossRef]
- Hunter, D.J.; Guermazi, A.; Lo, G.H.; Grainger, A.J.; Conaghan, P.G.; Boudreau, R.M.; Roemer, F.W. Evolution of Semi-Quantitative Whole Joint Assessment of Knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthr. Cartil. 2011, 19, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Madelin, G.; Poidevin, F.; Makrymallis, A.; Regatte, R.R. Classification of Sodium MRI Data of Cartilage Using Machine Learning. Magn. Reson. Med. 2015, 74, 1435–1448. [Google Scholar] [CrossRef]
- Nagel, A.M.; Laun, F.B.; Weber, M.A.; Matthies, C.; Semmler, W.; Schad, L.R. Sodium MRI Using a Density-Adapted 3D Radial Acquisition Technique. Magn. Reson. Med. 2009, 62, 1565–1573. [Google Scholar] [CrossRef]
- Haneder, S.; Konstandin, S.; Morelli, J.N.; Nagel, A.M.; Zoellner, F.G.; Schad, L.R.; Schoenberg, S.O.; Michaely, H.J. Quantitative and Qualitative 23 Na MR Imaging of the Human Kidneys at 3 T: Before and after a Water Load. Radiology 2011, 260, 857–865. [Google Scholar] [CrossRef]
- Cunningham, C.H.; Pauly, J.M.; Nayak, K.S. Saturated Double-Angle Method for Rapid B1+ Mapping. Magn. Reson. Med. 2006, 55, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Lommen, J.; Konstandin, S.; Krämer, P.; Schad, L.R. Enhancing the Quantification of Tissue Sodium Content by MRI: Time-Efficient Sodium B 1 Mapping at Clinical Field Strengths. NMR Biomed. 2016, 29, 129–136. [Google Scholar] [CrossRef]
- Müller-Lutz, A.; Kamp, B.; Nagel, A.M.; Ljimani, A.; Abrar, D.; Schleich, C.; Wollschläger, L.; Nebelung, S.; Wittsack, H.-J. Sodium MRI of Human Articular Cartilage of the Wrist: A Feasibility Study on a Clinical 3T MRI Scanner. Magn. Reson. Mater. Phys. Biol. Med. 2021, 34, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Madelin, G.; Lee, J.-S.; Inati, S.; Jerschow, A.; Regatte, R.R. Sodium Inversion Recovery MRI of the Knee Joint in Vivo at 7T. J. Magn. Reson. 2010, 207, 42–52. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wittsack, H.; Lanzman, R.S.; Mathys, C.; Janssen, H.; Mödder, U.; Blondin, D. Statistical Evaluation of Diffusion-weighted Imaging of the Human Kidney. Magn. Reson. Med. 2010, 64, 616–622. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A Reproducible Evaluation of ANTs Similarity Metric Performance in Brain Image Registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Wang, M.; Tsang, A.; Tam, V.; Chan, D.; Cao, P.; Wu, E.X. Multiparametric MR Investigation of Proteoglycan Diffusivity, T2 Relaxation, and Concentration in an Ex Vivo Model of Intervertebral Disc Degeneration. J. Magn. Reson. Imaging 2020, 51, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, A.; Shapiro, E.M.; Akella, S.V.S.; Gougoutas, A.; Kneeland, J.B.; Reddy, R. Quantifying Sodium in the Human Wrist in Vivo by Using MR Imaging. Radiology 2002, 224, 598–602. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Hafner, T.; Schock, J.; Post, M.; Abrar, D.B.; Sewerin, P.; Linka, K.; Knobe, M.; Kuhl, C.; Truhn, D.; Nebelung, S. A Serial Multiparametric Quantitative Magnetic Resonance Imaging Study to Assess Proteoglycan Depletion of Human Articular Cartilage and Its Effects on Functionality. Sci. Rep. 2020, 10, 15106. [Google Scholar] [CrossRef]
- Madelin, G.; Jerschow, A.; Regatte, R.R. Sodium Relaxation Times in the Knee Joint in Vivo at 7T. NMR Biomed. 2012, 25, 530–537. [Google Scholar] [CrossRef]
- Burstein, D.; Springer, C.S. Sodium MRI Revisited. Magn. Reson. Med. 2019, 82, 521–524. [Google Scholar] [CrossRef]
- Nagel, A.M.; Umathum, R.; Rösler, M.B.; Ladd, M.E.; Litvak, I.; Gor’kov, P.L.; Brey, W.W.; Schepkin, V.D. 39 K and 23 Na Relaxation Times and MRI of Rat Head at 21.1 T. NMR Biomed. 2016, 29, 759–766. [Google Scholar] [CrossRef]
- Borthakur, A.; Hancu, I.; Boada, F.E.; Shen, G.X.; Shapiro, E.M.; Reddy, R. In Vivo Triple Quantum Filtered Twisted Projection Sodium MRI of Human Articular Cartilage. J. Magn. Reson. 1999, 141, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Zaric, O.; Juras, V.; Szomolanyi, P.; Schreiner, M.; Raudner, M.; Giraudo, C.; Trattnig, S. Frontiers of Sodium MRI Revisited: From Cartilage to Brain Imaging. J. Magn. Reson. Imaging 2021, 54, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Bangerter, N.K.; Kaggie, J.D.; Taylor, M.D.; Hadley, J.R. Sodium MRI Radiofrequency Coils for Body Imaging. NMR Biomed. 2016, 29, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Lachner, S.; Utzschneider, M.; Zaric, O.; Minarikova, L.; Ruck, L.; Zbýň, Š.; Hensel, B.; Trattnig, S.; Uder, M.; Nagel, A.M. Compressed Sensing and the Use of Phased Array Coils in 23Na MRI: A Comparison of a SENSE-Based and an Individually Combined Multi-Channel Reconstruction. Z. Med. Phys. 2021, 31, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Madelin, G.; Chang, G.; Otazo, R.; Jerschow, A.; Regatte, R.R. Compressed Sensing Sodium MRI of Cartilage at 7T: Preliminary Study. J. Magn. Reson. 2012, 214, 360–365. [Google Scholar] [CrossRef]
- Kratzer, F.J.; Flassbeck, S.; Schmitter, S.; Wilferth, T.; Magill, A.W.; Knowles, B.R.; Platt, T.; Bachert, P.; Ladd, M.E.; Nagel, A.M. 3D Sodium (23 Na) Magnetic Resonance Fingerprinting for Time-efficient Relaxometric Mapping. Magn. Reson. Med. 2021, 86, 2412–2425. [Google Scholar] [CrossRef] [PubMed]
- Khajehim, M.; Christen, T.; Tam, F.; Graham, S.J. Streamlined Magnetic Resonance Fingerprinting: Fast Whole-Brain Coverage with Deep-Learning Based Parameter Estimation. Neuroimage 2021, 238, 118237. [Google Scholar] [CrossRef] [PubMed]
- Müller-Lutz, A.; Schleich, C.; Pentang, G.; Schmitt, B.; Lanzman, R.S.; Matuschke, F.; Wittsack, H.-J.; Miese, F. Age-Dependency of Glycosaminoglycan Content in Lumbar Discs: A 3t GagcEST Study. J. Magn. Reson. Imaging 2015, 42, 1517–1523. [Google Scholar] [CrossRef]
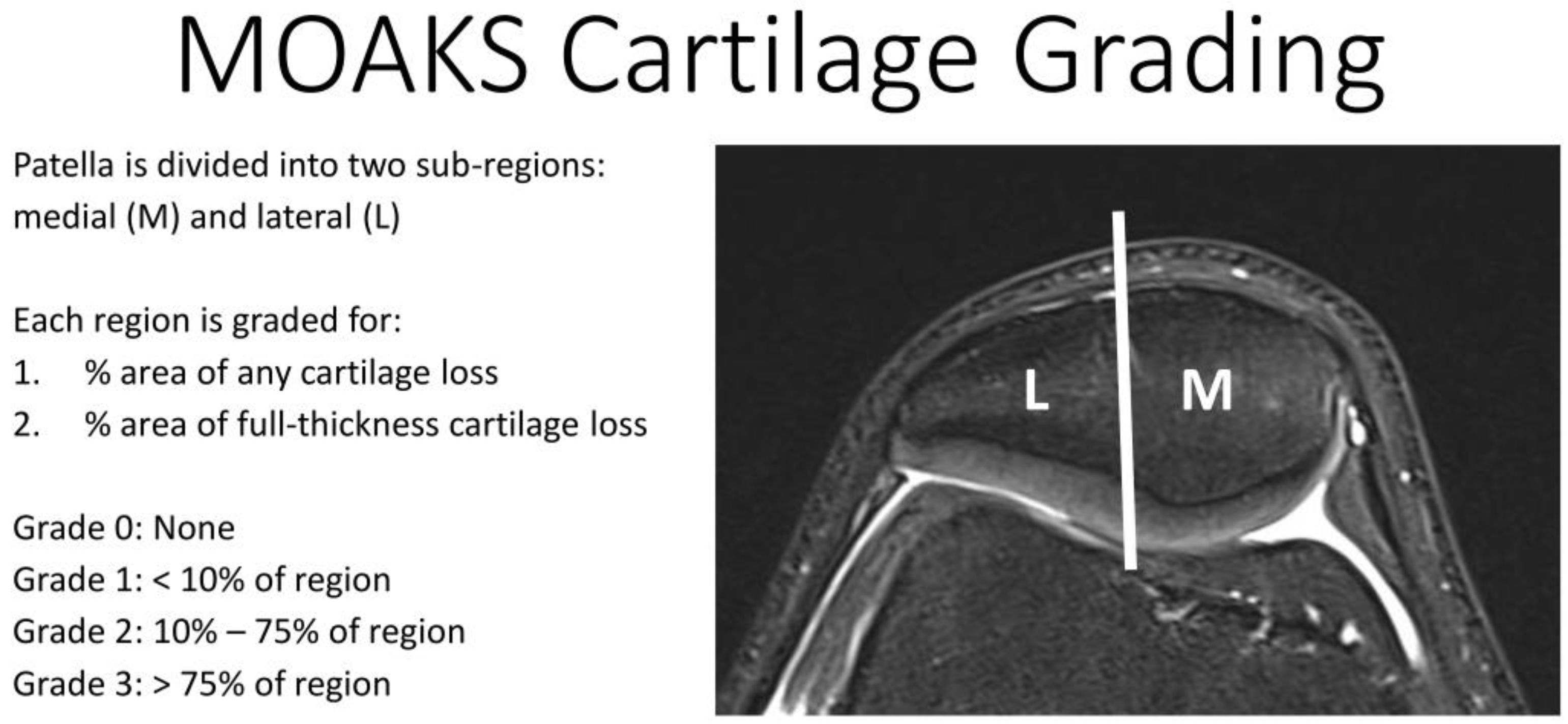

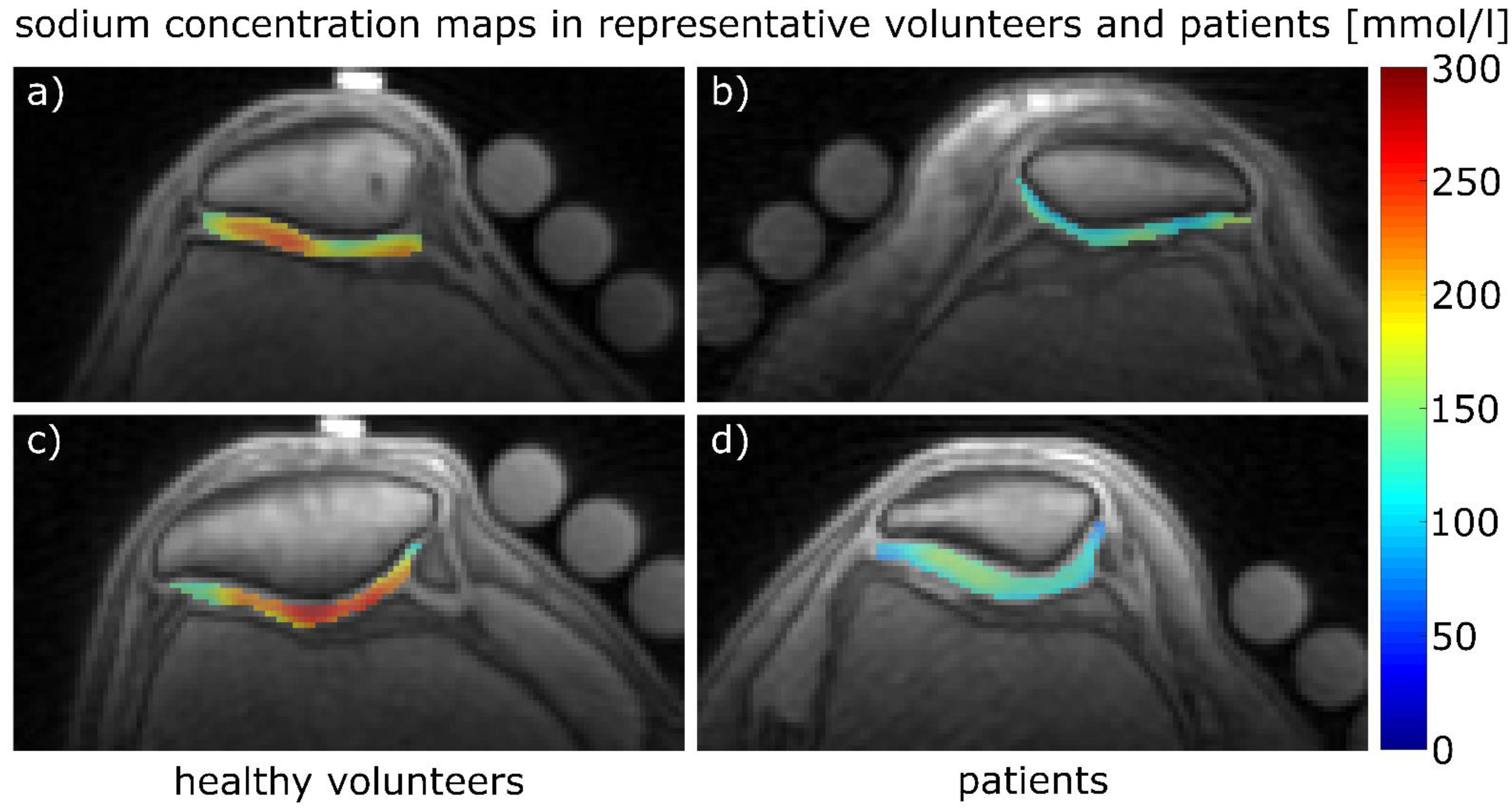
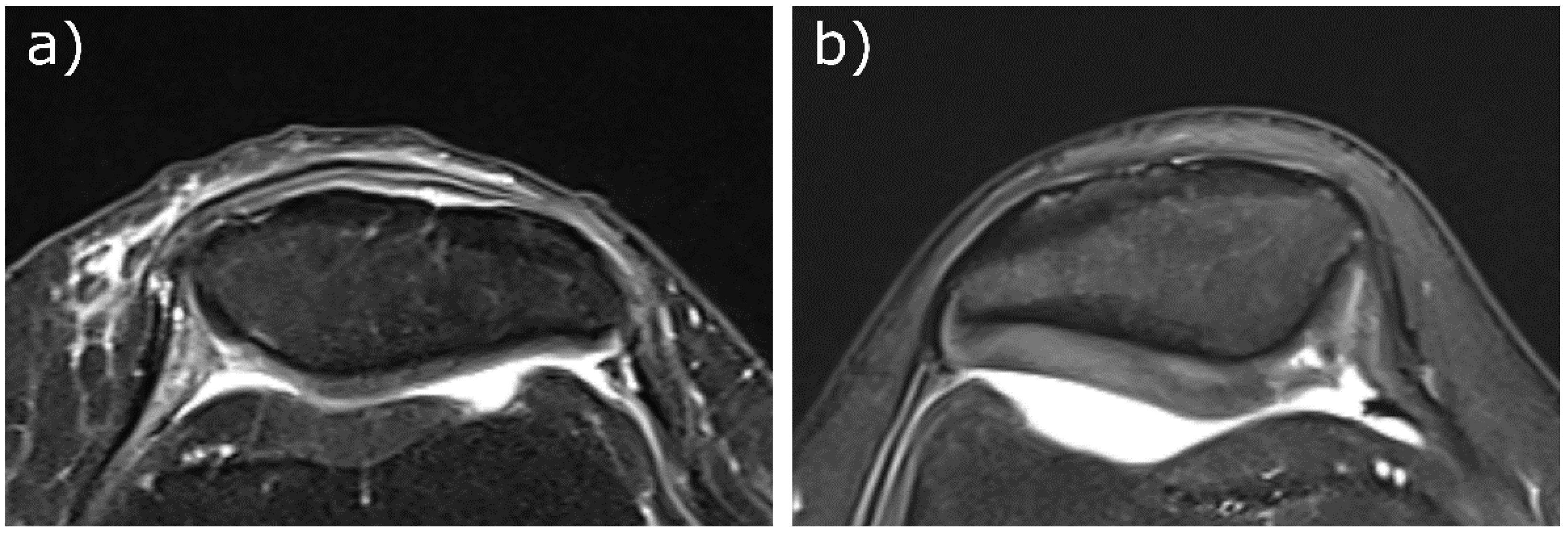
| 23Na Coil Sensitivity | B1 Mapping | Protocol 1 | Protocol 2 | 1H Imaging | |
|---|---|---|---|---|---|
| Sequence type | DA-3D-RAD | DA-3D-RAD | DA-3D-RAD | DA-3D-RAD | DA-3D-RAD |
| Nucleus | 23Na | 23Na | 23Na | 23Na | 1H |
| Orientation | tra | tra | tra | tra | tra |
| Repetition time (ms) | 60 | 300 | 8/9/10/11/12/13/14/15/16/18/20/23/26/30/40/50/70 | 84 | 30 |
| Echo time (ms) | 0.3 | 0.3 | 0.3 | (0.30/6.45/12.60/18.80) (1.50/7.65/13.80/20.00) (3.00/9.15/15.30/21.50) | 0.8 |
| Inversion time (ms) | - | - | - | 24 | - |
| Inversion pulse Duration (ms) | - | - | - | 1 | - |
| Field of View (mm) | 180 × 180 × 180 | 180 × 180 × 180 | 180 × 180 × 180 | 180 × 180 × 180 | 180 × 180 × 180 |
| Projections | 50,000 | 50,000 | 9000 | 9000 | 9000 |
| Pixel size (mm/px) | 3 × 3 × 3 | 3 × 3 × 3 | 3 × 3 × 3 | 3 × 3 × 3 | 1 × 1 × 1 |
| Flip angle (°) | 90 | 40/80 | 90 | 90 | 10 |
| Pulse duration (ms) | 0.5 | 0.5 | 0.5 | 0.5 | 0.2 |
| Readout time (ms) | 5 | 5 | 5 | 5 | 1 |
| Averages | 12 | 2 | 1 | 1 | 1 |
| Total examination time (h:min:s) | 10:00:00 | 16:40:00 | 00:57:45 | 00:37:48 | 00:04:30 |
| PD-Weighted fs | ||
|---|---|---|
| Sequence type | TSE | TSE |
| Turbo Factor | 38 | 109 |
| GRAPPA | 2 | 2 |
| Orientation | cor/tra/sag | sag |
| Repetition time (ms) | 4980 | 864 |
| Echo time (ms) | 42 | 13 |
| Field of View (mm) | 160 × 160 | 160 × 160 |
| Image matrix (px) | 512 × 512 | 512 × 512 |
| Pixel size (mm/px) | 0.3 × 0.3 | 0.3 × 0.3 |
| Flip angle (°) | 180 | 180 |
| Slices | 35 | 35 |
| Slice thickness (mm) | 3 | 3 |
| Total examination time (min:s) | 09:57 | 03:10 |
| Radiologist/ | Volunteers | Patient | |||||
|---|---|---|---|---|---|---|---|
| Measurement | Mean | Std | Min | Median | Max | ||
| 1/1 | 14.45 | 0.74 | 13.24 | 14.47 | 15.33 | 15.42 | |
| (ms) | 1/2 | 14.58 | 0.74 | 13.24 | 14.78 | 15.36 | 15.60 |
| 2/1 | 14.61 | 0.68 | 13.26 | 14.80 | 15.29 | 15.60 | |
| 1/1 | 37.91 | 2.92 | 35.07 | 37.79 | 44.24 | 39.78 | |
| (ms) | 1/2 | 38.32 | 2.89 | 35.02 | 39.24 | 44.50 | 39.65 |
| 2/1 | 38.88 | 2.86 | 35.07 | 39.24 | 43.81 | 39.99 | |
| 1/1 | 77.30 | 3.73 | 72.80 | 78.53 | 82.64 | 71.23 | |
| (%) | 1/2 | 76.54 | 3.73 | 72.71 | 75.34 | 82.60 | 71.03 |
| 2/1 | 75.10 | 4.15 | 68.10 | 73.58 | 82.55 | 70.69 | |
| 1/1 | 0.9917 | 0.0045 | 0.9808 | 0.9926 | 0.9959 | 0.9829 | |
| 1/2 | 0.9918 | 0.0040 | 0.9831 | 0.9932 | 0.9962 | 0.9907 | |
| 2/1 | 0.9918 | 0.0045 | 0.9806 | 0.9930 | 0.9971 | 0.9903 |
| Radiologist/ | Volunteers | Patient | |||||
|---|---|---|---|---|---|---|---|
| Measurement | Mean | Std | Min | Median | Max | ||
| 1/1 | 0.358 | 0.147 | 0.103 | 0.353 | 0.663 | 0.105 | |
| (ms) | 1/2 | 0.365 | 0.155 | 0.104 | 0.370 | 0.677 | 0.107 |
| 2/1 | 0.365 | 0.174 | 0.105 | 0.357 | 0.751 | 0.107 | |
| 1/1 | 12.62 | 0.73 | 11.30 | 12.55 | 13.74 | 13.99 | |
| (ms) | 1/2 | 12.77 | 0.70 | 11.35 | 12.78 | 13.74 | 14.00 |
| 2/1 | 12.79 | 0.72 | 11.49 | 12.71 | 13.98 | 14.00 | |
| 1/1 | 34.39 | 4.78 | 25.35 | 34.48 | 41.44 | 25.86 | |
| (%) | 1/2 | 34.11 | 4.92 | 25.37 | 34.28 | 41.72 | 26.00 |
| 2/1 | 33.81 | 5.09 | 25.28 | 34.66 | 41.79 | 25.99 | |
| 1/1 | 0.9856 | 0.0088 | 0.9643 | 0.9879 | 0.9931 | 0.9623 | |
| R2 | 1/2 | 0.9870 | 0.0097 | 0.9631 | 0.9910 | 0.9960 | 0.9657 |
| 2/1 | 0.9861 | 0.0093 | 0.9640 | 0.9888 | 0.9949 | 0.9650 |
| Radiologist/ | Volunteers | Patient | ||||||
|---|---|---|---|---|---|---|---|---|
| Measurement | Mean | Std | Min | Median | Max | p-Value | ||
| Protocol 1 | 1/1 | 215 | 44 | 166 | 203 | 291 | 0.441 | 135 |
| 23Na-Conc. | 1/2 | 204 | 40 | 169 | 187 | 276 | 0.859 | 129 |
| (mmol/L) | 2/1 | 218 | 52 | 151 | 203 | 297 | 0.374 | 136 |
| Protocol 2 | 1/1 | 200 | 48 | 130 | 199 | 267 | - | 158 |
| 23Na-Conc. | 1/2 | 194 | 45 | 134 | 188 | 261 | - | 152 |
| (mmol/L) | 2/1 | 204 | 39 | 134 | 201 | 291 | - | 136 |
| Magnetic Field Strength of MRI Scanner (T) | 23Na Relaxation Time Results for Patellar Cartilage and Synovial Fluid | |||||
|---|---|---|---|---|---|---|
| Madelin et al. [39] | 7.0 | 17.7 ± 2.6 | - | 0.5 ± 0.1 | 11.4 ± 1.8 | 39 ± 4 |
| Feldman et al. [20] | 4.7 | 21 ± 1 | 48 ± 3 | 0.8 ± 0.2 | 19.7 ± 0.5 | 65 ± 12 |
| Staroswiecki et al. [21] | 7.0 | - | - | - | 13.2 ± 1.5 | - |
| 3.0 | - | - | - | 15.5 ± 1.3 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamp, B.; Frenken, M.; Henke, J.M.; Abrar, D.B.; Nagel, A.M.; Gast, L.V.; Oeltzschner, G.; Wilms, L.M.; Nebelung, S.; Antoch, G.; et al. Quantification of Sodium Relaxation Times and Concentrations as Surrogates of Proteoglycan Content of Patellar CARTILAGE at 3T MRI. Diagnostics 2021, 11, 2301. https://doi.org/10.3390/diagnostics11122301
Kamp B, Frenken M, Henke JM, Abrar DB, Nagel AM, Gast LV, Oeltzschner G, Wilms LM, Nebelung S, Antoch G, et al. Quantification of Sodium Relaxation Times and Concentrations as Surrogates of Proteoglycan Content of Patellar CARTILAGE at 3T MRI. Diagnostics. 2021; 11(12):2301. https://doi.org/10.3390/diagnostics11122301
Chicago/Turabian StyleKamp, Benedikt, Miriam Frenken, Jan M. Henke, Daniel B. Abrar, Armin M. Nagel, Lena V. Gast, Georg Oeltzschner, Lena M. Wilms, Sven Nebelung, Gerald Antoch, and et al. 2021. "Quantification of Sodium Relaxation Times and Concentrations as Surrogates of Proteoglycan Content of Patellar CARTILAGE at 3T MRI" Diagnostics 11, no. 12: 2301. https://doi.org/10.3390/diagnostics11122301
APA StyleKamp, B., Frenken, M., Henke, J. M., Abrar, D. B., Nagel, A. M., Gast, L. V., Oeltzschner, G., Wilms, L. M., Nebelung, S., Antoch, G., Wittsack, H.-J., & Müller-Lutz, A. (2021). Quantification of Sodium Relaxation Times and Concentrations as Surrogates of Proteoglycan Content of Patellar CARTILAGE at 3T MRI. Diagnostics, 11(12), 2301. https://doi.org/10.3390/diagnostics11122301







