Predicting Prolonged Length of ICU Stay through Machine Learning
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Datasets and Study Subjects
3.2. Primary Outcome and Predictor Variables
3.3. Model Development
3.3.1. Support Vector Machine (SVM)
3.3.2. Random Forest (RF)
3.3.3. Gradient Boosting Decision Tree (GBDT)
3.3.4. Deep Learning (DL)
3.3.5. Customized SAPS II
3.4. Model Validation
3.5. Analysis
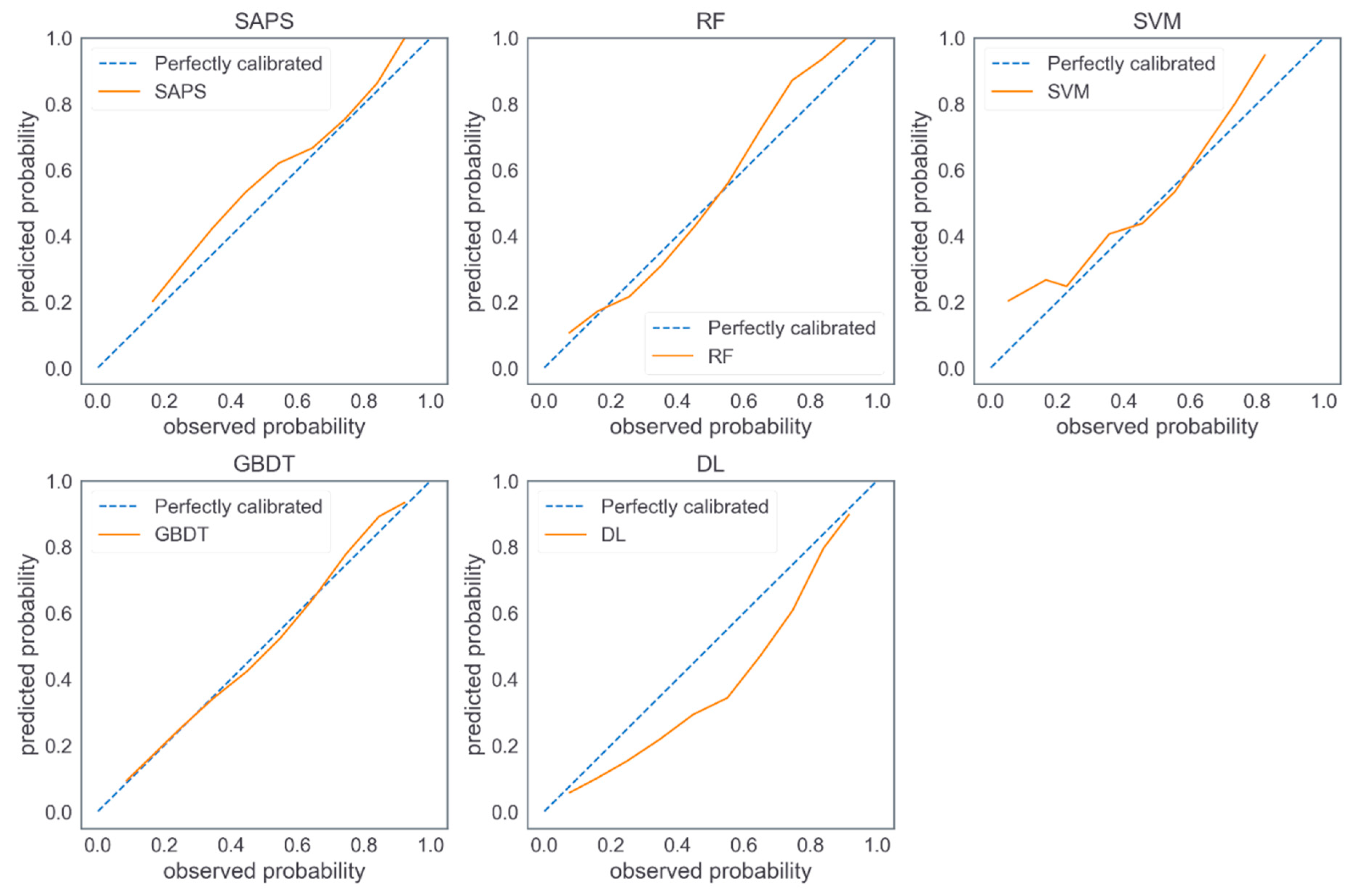
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyer, A.; Zverinski, D.; Pfahringer, B.; Kempfert, J.; Kuehne, T.; Sündermann, S.H.; Stamm, C.; Hofmann, T.; Falk, V.; Eickhoff, C. Machine learning for real-time prediction of complications in critical care: A retrospective study. Lancet Respir. Med. 2018, 6, 905–914. [Google Scholar] [CrossRef]
- Vincent, J.L.; Marshall, J.C.; Namendys-Silva, S.A.; Francois, B.; Martin-Loeches, I.; Lipman, J.; Reinhart, K.; Antonelli, M.; Pickkers, P.; Njimi, H.; et al. Assessment of the worldwide burden of critical illness: The Intensive Care Over Nations (ICON) audit. Lancet Resp. Med. 2014, 2, 380–386. [Google Scholar] [CrossRef]
- Halpern, N.A.; Pastores, S.M. Critical care medicine in the United States 2000-2005: An analysis of bed numbers, occupancy rates, payer mix, and costs. Crit. Care Med. 2010, 38, 65–71. [Google Scholar] [CrossRef]
- Verburg, I.W.M.; Atashi, A.; Eslami, S.; Holman, R.; Abu-Hanna, A.; de Jonge, E.; Peek, N.; de Keizer, N.F. Which models can I use to predict adult ICU length of stay? A systematic review. Crit. Care Med. 2017, 45, e222–e231. [Google Scholar] [CrossRef] [PubMed]
- Kılıç, M.; Yüzkat, N.; Soyalp, C.; Gülhaş, N. Cost analysis on Intensive Care Unit costs based on the length of stay. Turk. J. Anaesthesiol. Reanim. 2019, 47, 142. [Google Scholar] [CrossRef]
- Herman, C.; Karolak, W.; Yip, A.M.; Buth, K.J.; Hassan, A.; Legare, J.-F. Predicting prolonged intensive care unit length of stay in patients undergoing coronary artery bypass surgery—Development of an entirely preoperative scorecard. Interact. Cardiovasc. Thorac. Surg. 2009, 9, 654–658. [Google Scholar] [CrossRef]
- Verburg, I.W.M.; de Keizer, N.F.; de Jonge, E.; Peek, N. Comparison of Regression Methods for Modeling Intensive Care Length of Stay. PLoS ONE 2014, 9, e109684. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.; Kobewka, D.; Thavorn, K.; D’Egidio, G.; Rosenberg, E.; Kyeremanteng, K. The impact of reducing intensive care unit length of stay on hospital costs: Evidence from a tertiary care hospital in Canada. Can. J. Anesth. J. Can. d’anesthésie 2018, 65, 627–635. [Google Scholar] [CrossRef]
- Stricker, K.; Rothen, H.U.; Takala, J. Resource use in the ICU: Short- vs. long-term patients. Acta Anaesthesiol. Scand. 2003, 47, 508–515. [Google Scholar] [CrossRef]
- Nassar, A.P.; Caruso, P. ICU physicians are unable to accurately predict length of stay at admission: A prospective study. Int. J. Qual. Health Care 2016, 28, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.; Meloni, F.; Baranauskas, J.A.; Macedo, A.A. Prediction of mortality in Intensive Care Units: A multivariate feature selection. J. Biomed. Inform. 2020, 107, 103456. [Google Scholar] [CrossRef]
- Pereira, T.; Gadhoumi, K.; Ma, M.; Liu, X.; Xiao, R.; Colorado, R.A.; Keenan, K.J.; Meisel, K.; Hu, X. A Supervised Approach to Robust Photoplethysmography Quality Assessment. IEEE J. Biomed. Health Inform. 2020, 24, 649–657. [Google Scholar] [CrossRef]
- Qian, Q.; Wu, J.; Wang, J.; Sun, H.; Yang, L. Prediction Models for AKI in ICU: A Comparative Study. Int. J. Gen. Med. 2021, 14, 623–632. [Google Scholar] [CrossRef]
- Li, X.; Ge, P.; Zhu, J.; Li, H.; Graham, J.; Singer, A.; Richman, P.S.; Duong, T.Q. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ 2020, 8, e10337. [Google Scholar] [CrossRef]
- Theis, J.; Galanter, W.; Boyd, A.; Darabi, H. Improving the In-Hospital Mortality Prediction of Diabetes ICU Patients Using a Process Mining/Deep Learning Architecture. IEEE J. Biomed. Health Inform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zoller, B.; Spanaus, K.; Gerster, R.; Fasshauer, M.; Stehberger, P.A.; Klinzing, S.; Vergopoulos, A.; von Eckardstein, A.; Bechir, M. ICG-liver test versus new biomarkers as prognostic markers for prolonged length of stay in critically ill patients—A prospective study of accuracy for prediction of length of stay in the ICU. Ann. Intensive Care 2014, 4, 5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eachempati, S.R.; Hydo, L.J.; Barie, P.S. Severity scoring for prognostication in patients with severe acute pancreatitis—Comparative analysis of the ranson score and the APACHE III score. Arch. Surg. 2002, 137, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Houthooft, R.; Ruyssinck, J.; van der Herten, J.; Stijven, S.; Couckuyt, I.; Gadeyne, B.; Ongenae, F.; Colpaert, K.; Decruyenaere, J.; Dhaene, T.; et al. Predictive modelling of survival and length of stay in critically ill patients using sequential organ failure scores. Artif. Intell. Med. 2015, 63, 191–207. [Google Scholar] [CrossRef]
- Rotar, E.P.; Beller, J.P.; Smolkin, M.E.; Chancellor, W.Z.; Ailawadi, G.; Yarboro, L.T.; Hulse, M.; Ratcliffe, S.J.; Teman, N.R. Prediction of prolonged intensive care unit length of stay following cardiac surgery. In Seminars in Thoracic and Cardiovascular Surgery; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Lin, K.; Hu, Y.; Kong, G. Predicting in-hospital mortality of patients with acute kidney injury in the ICU using random forest model. Int. J. Med Inform. 2019, 125, 55–61. [Google Scholar] [CrossRef]
- Wellner, B.; Grand, J.; Canzone, E.; Coarr, M.; Brady, P.W.; Simmons, J.; Kirkendall, E.; Dean, N.; Kleinman, M.; Sylvester, P. Predicting Unplanned Transfers to the Intensive Care Unit: A Machine Learning Approach Leveraging Diverse Clinical Elements. JMIR Med. Inf. 2017, 5, 16. [Google Scholar] [CrossRef]
- Pirracchio, R.; Petersen, M.L.; Carone, M.; Rigon, M.R.; Chevret, S.; van der Laan, M.J. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Resp. Med. 2015, 3, 42–52. [Google Scholar] [CrossRef]
- Gonzalez-Robledo, J.; Martin-Gonzalez, F.; Sanchez-Barba, M.; Sanchez-Hernandez, F.; Moreno-Garcia, M.N. Multiclassifier Systems for Predicting Neurological Outcome of Patients with Severe Trauma and Polytrauma in Intensive Care Units. J. Med. Syst. 2017, 41, 8. [Google Scholar] [CrossRef]
- Meiring, C.; Dixit, A.; Harris, S.; MacCallum, N.S.; Brealey, D.A.; Watkinson, P.J.; Jones, A.; Ashworth, S.; Beale, R.; Brett, S.J.; et al. Optimal intensive care outcome prediction over time using machine learning. PLoS ONE 2018, 13, e0206862. [Google Scholar] [CrossRef] [PubMed]
- Viton, F.; Elbattah, M.; Guérin, J.-L.; Dequen, G. Multi-channel ConvNet Approach to Predict the Risk of in-Hospital Mortality for ICU Patients. DeLTA 2020, 98–102. [Google Scholar]
- Navaz, A.N.; Mohammed, E.; Serhani, M.A.; Zaki, N. The Use of Data Mining Techniques to Predict Mortality and Length of Stay in an ICU. In Proceedings of the 2016 12th International Conference on Innovations in Information Technology, Abu Dhabi, United Arab Emirates, 28–30 November 2016; IEEE: New York, NY, USA, 2016; pp. 164–168. [Google Scholar]
- Rocheteau, E.; Liò, P.; Hyland, S. Temporal pointwise convolutional networks for length of stay prediction in the intensive care unit. In Proceedings of the Conference on Health, Inference, and Learning, Virtual Event, Association for Computing Machinery, New York, NY, USA, 8–10 April 2021; pp. 58–68. [Google Scholar]
- Ma, X.; Si, Y.; Wang, Z.; Wang, Y. Length of stay prediction for ICU patients using individualized single classification algorithm. Comput. Methods Programs Biomed. 2020, 186, 105224. [Google Scholar] [CrossRef] [PubMed]
- Vasilevskis, E.E.; Kuzniewicz, M.W.; Cason, B.A.; Lane, R.K.; Dean, M.L.; Clay, T.; Rennie, D.J.; Vittinghoff, E.; Dudley, R.A. Mortality Probability Model III and Simplified Acute Physiology Score II Assessing Their Value in Predicting Length of Stay and Comparison to APACHE IV. Chest 2009, 136, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Ettema, R.G.A.; Peelen, L.M.; Schuurmans, M.J.; Nierich, A.P.; Kalkman, C.J.; Moons, K.G.M. Prediction Models for Prolonged Intensive Care Unit Stay After Cardiac Surgery Systematic Review and Validation Study. Circulation 2010, 122, 682–689. [Google Scholar] [CrossRef]
- Pollard, T.J.; Johnson, A.E.W.; Raffa, J.D.; Celi, L.A.; Mark, R.G.; Badawi, O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci. Data 2018, 5, 13. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.W.H.; Feng, M.L.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Soveri, I.; Snyder, J.; Holdaas, H.; Holme, I.; Jardine, A.G.; L’Italien, G.J.; Fellstrom, B. The External Validation of the Cardiovascular Risk Equation for Renal Transplant Recipients: Applications to BENEFIT and BENEFIT-EXT Trials. Transplantation 2013, 95, 142–147. [Google Scholar] [CrossRef]
- Grooms, K.N.; Ommerborn, M.J.; Pham, D.Q.; Djousse, L.; Clark, C.R. Dietary Fiber Intake and Cardiometabolic Risks among US Adults, NHANES 1999-2010. Am. J. Med. 2013, 126, 1059–1067.e4. [Google Scholar] [CrossRef] [PubMed]
- Harel, Z.; Wald, R.; McArthur, E.; Chertow, G.M.; Harel, S.; Gruneir, A.; Fischer, N.D.; Garg, A.X.; Perl, J.; Nash, D.M.; et al. Rehospitalizations and Emergency Department Visits after Hospital Discharge in Patients Receiving Maintenance Hemodialysis. J. Am. Soc. Nephrol. 2015, 26, 3141–3150. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xia, C.; Li, Q.; Yao, C.; Yao, Y.; Chen, D.; Jiang, Q. Perioperative predictors of prolonged length of hospital stay following total knee arthroplasty: A retrospective study from a single center in China. BMC Musculoskelet. Disord. 2020, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Lilly, C.M.; Zuckerman, I.H.; Badawi, O.; Riker, R.R. Benchmark Data From More Than 240,000 Adults That Reflect the Current Practice of Critical Care in the United States. Chest 2011, 140, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Legall, J.R.; Lemeshow, S.; Saulnier, F. A new simplified acute physiology score (SAPS-II) based on a European North-American multicenter study. JAMA 1993, 270, 2957–2963. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Zoppis, I.; Mauri, G.; Dondi, R. Kernel methods: Support vector machines. Encycl. Bioinform. Comput. Biol. 2019, 1, 503–510. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Fayed, H.A.; Atiya, A.F. Speed up grid-search for parameter selection of support vector machines. Appl. Soft Comput. 2019, 80, 202–210. [Google Scholar] [CrossRef]
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef]
- Supatcha, L.; Chinae, T.; Chakarida, N.; Boonserm, K.; Marasri, R. Heterogeneous ensemble approach with discriminative features and modified-SMOTEbagging for pre-miRNA classification. Nucleic Acids Res. 2013, 41, e21. [Google Scholar]
- Kulkarni, V.Y.; Sinha, P.K.; Petare, M.C. Weighted Hybrid Decision Tree Model for Random Forest Classifier. J. Inst. Eng. 2016, 97, 209–217. [Google Scholar] [CrossRef]
- Luo, R.; Tan, X.; Wang, R.; Qin, T.; Chen, E.; Liu, T.-Y. Accuracy Prediction with Non-neural Model for Neural Architecture Search. arXiv 2020, arXiv:2007.04785. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, JMLR Workshop and Conference Proceedings, Montreal, QC, Canada, 31 March 2010; pp. 249–256. [Google Scholar]
- Yu, H.; Liu, J.; Chen, C.; Heidari, A.A.; Zhang, Q.; Chen, H.; Mafarja, M.; Turabieh, H. Corn leaf diseases diagnosis based on K-means clustering and deep learning. IEEE Access 2021, 9, 143824–143835. [Google Scholar] [CrossRef]
- Gu, J.; Tong, T.; He, C.; Xu, M.; Yang, X.; Tian, J.; Jiang, T.; Wang, K. Deep learning radiomics of ultrasonography can predict response to neoadjuvant chemotherapy in breast cancer at an early stage of treatment: A prospective study. Eur. Radiol. 2021, 1–11. [Google Scholar] [CrossRef]
- Yu, K.; Yang, Z.; Wu, C.; Huang, Y.; Xie, X. In-hospital resource utilization prediction from electronic medical records with deep learning. Knowl.-Based Syst. 2021, 223, 107052. [Google Scholar] [CrossRef]
- Imai, S.; Takekuma, Y.; Kashiwagi, H.; Miyai, T.; Kobayashi, M.; Iseki, K.; Sugawara, M. Validation of the usefulness of artificial neural networks for risk prediction of adverse drug reactions used for individual patients in clinical practice. PLoS ONE 2020, 15, e0236789. [Google Scholar] [CrossRef]
- Zeng, Z.; Yao, S.; Zheng, J.; Gong, X. Development and validation of a novel blending machine learning model for hospital mortality prediction in ICU patients with Sepsis. BioData Min. 2021, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Dzaharudin, F.; Ralib, A.; Jamaludin, U.; Nor, M.; Tumian, A.; Har, L.; Ceng, T. Mortality Prediction in Critically Ill Patients Using Machine Learning Score; IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2020. [Google Scholar]
- Fallenius, M.; Skrifvars, M.B.; Reinikainen, M.; Bendel, S.; Raj, R. Common intensive care scoring systems do not outperform age and glasgow coma scale score in predicting mid-term mortality in patients with spontaneous intracerebral hemorrhage treated in the intensive care unit. Scand. J. Trauma Resusc. Emerg. Med. 2017, 25, 102. [Google Scholar] [CrossRef]
- Jahn, M.; Rekowski, J.; Gerken, G.; Kribben, A.; Canbay, A.; Katsounas, A. The predictive performance of SAPS 2 and SAPS 3 in an intermediate care unit for internal medicine at a German university transplant center; A retrospective analysis. PLoS ONE 2019, 14, e0222164. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the Performance of Prediction Models A Framework for Traditional and Novel Measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Van Hoorde, K.; Van Huffel, S.; Timmerman, D.; Bourne, T.; Van Calster, B. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. J. Biomed. Inform. 2015, 54, 283–293. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar]
- Wang, S.Q.; Yang, J.; Chou, K.C. Using stacked generalization to predict membrane protein types based on pseudo-amino acid composition. J. Theor. Biol. 2006, 242, 941–946. [Google Scholar] [CrossRef]
- Phan, J.H.; Hoffman, R.; Kothari, S.; Wu, P.Y.; Wang, M.D. Integration of Multi-Modal Biomedical Data to Predict Cancer Grade and Patient Survival; IEEE: New York, NY, USA, 2016; pp. 577–580. [Google Scholar]
- Zhang, C.S.; Liu, C.C.; Zhang, X.L.; Almpanidis, G. An up-to-date comparison of state-of-the-art classification algorithms. Expert Syst. Appl. 2017, 82, 128–150. [Google Scholar] [CrossRef]
- Ichikawa, D.; Saito, T.; Ujita, W.; Oyama, H. How can machine-learning methods assist in virtual screening for hyperuricemia? A healthcare machine-learning approach. J. Biomed. Inform. 2016, 64, 20–24. [Google Scholar] [CrossRef]
- Zhou, C.; Yu, H.; Ding, Y.J.; Guo, F.; Gong, X.J. Multi-scale encoding of amino acid sequences for predicting protein interactions using gradient boosting decision tree. PLoS ONE 2017, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Hotzy, F.; Theodoridou, A.; Hoff, P.; Schneeberger, A.R.; Seifritz, E.; Olbrich, S.; Jager, M. Machine Learning: An Approach in Identifying Risk Factors for Coercion Compared to Binary Logistic Regression. Front. Psychiatry 2018, 9, 11. [Google Scholar] [CrossRef]
- Xiong, J. Radial Distance Weighted Discrimination. Ph.D. Thesis, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; 120p.
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2021, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Mehta, R.; Chinthapalli, K. Glasgow coma scale explained. BMJ 2019, 365, l1296. [Google Scholar] [CrossRef] [PubMed]
- Al-Mufti, F.; Amuluru, K.; Lander, M.; Mathew, M.; El-Ghanem, M.; Nuoman, R.; Park, S.; Patel, V.; Singh, I.P.; Gupta, G. Low glasgow coma score in traumatic intracranial hemorrhage predicts development of cerebral vasospasm. World Neurosurg. 2018, 120, e68–e71. [Google Scholar] [CrossRef] [PubMed]
- Ghelichkhani, P.; Esmaeili, M.; Hosseini, M.; Seylani, K. Glasgow coma scale and four score in predicting the mortality of trauma patients; a diagnostic accuracy study. Emergency 2018, 6, e42. [Google Scholar]
- Gayat, E.; Cariou, A.; Deye, N.; Vieillard-Baron, A.; Jaber, S.; Damoisel, C.; Lu, Q.; Monnet, X.; Rennuit, I.; Azoulay, E. Determinants of long-term outcome in ICU survivors: Results from the FROG-ICU study. Crit. Care 2018, 22, 8. [Google Scholar] [CrossRef] [PubMed]
- Sahin, S.; Dogan, U.; Ozdemir, K.; Gok, H. Evaluation of clinical and demographic characteristics and their association with length of hospital stay in patients admitted to cardiac intensive care unit with the diagnosis of acute heart failure. Anat. J. Cardiol. 2012, 12, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Esteve, F.; Lopez-Delgado, J.C.; Javierre, C.; Skaltsa, K.; Carrio, M.L.L.; Rodriguez-Castro, D.; Torrado, H.; Farrero, E.; Diaz-Prieto, A.; Ventura, J.L.L.; et al. Evaluation of the PaO2/FiO2 ratio after cardiac surgery as a predictor of outcome during hospital stay. BMC Anesthesiol. 2014, 14, 9. [Google Scholar] [CrossRef]
- Piriyapatsom, A.; Williams, E.C.; Waak, K.; Ladha, K.S.; Eikermann, M.; Schmidt, U.H. Prospective Observational Study of Predictors of Re-Intubation Following Extubation in the Surgical ICU. Respir. Care 2016, 61, 306–315. [Google Scholar] [CrossRef]
- Faisst, M.; Wellner, U.F.; Utzolino, S.; Hopt, U.T.; Keck, T. Elevated blood urea nitrogen is an independent risk factor of prolonged intensive care unit stay due to acute necrotizing pancreatitis. J. Crit. Care 2010, 25, 105–111. [Google Scholar] [CrossRef]

| Category | Study | Population | Sample Size | Dataset | Outcome | Models | External Validation | Performance |
|---|---|---|---|---|---|---|---|---|
| Traditional regression based pLOS-ICU prediction models | Zoller et al. [16] | General ICU patients | 110 | Single-center | pLOS-ICU | Customized SAPS II | × | AUROC: 0.70 |
| Houthooft et al. [18] | General ICU patients | 14,480 | Single-center | pLOS-ICU | Customized SOFA score | × | Sensitivity: 0.71 | |
| Herman et al. [6] | Patients undergoing CABG | 3483 | Single-center | pLOS-ICU | LR | × | AUROC: 0.78 | |
| Rotar et al. [19] | Patients following CABG | 3283 | Single-center | pLOS-ICU | LASSO | × | AUROC: 0.72 | |
| ML-based models in ICU | Meiring et al. [24] | General ICU patients | 22,514 | Multicenter | ICU mortality | AdaBoost, RF, SVM, DL, LR, and customized APACHE-II | × | AUROC: 0.88 (DL) |
| Lin et al. [20] | Acute kidney injury patients | 19,044 | Single-center | ICU mortality | ANN, SVM, RF, and customized SAPS II | × | AUROC: 0.87 (RF) | |
| Viton et al. [25] | General ICU patients | 13,000 | Single-center | ICU mortality | DL | × | AUROC: 0.85 | |
| Qian et al. [13] | General ICU patients | 17,205 | Single-center | Acute kidney injury | XGBoost, RF, SVM, GBDT, DL, and LR | × | AUROC: 0.91 (GBDT) | |
| ML-based pLOS-ICU prediction models | Navaz et al. [26] | General ICU patients | 40,426 | Single-center | pLOS-ICU | Decision tree | × | Accuracy: 0.59 |
| Rocheteau et al. [27] | General ICU patients | 168,577 | Multicenter | LOS-ICU | DL | √ | R2: 0.40 | |
| Ma et al. [28] | General ICU patients | 4000 | Single-center | pLOS-ICU | Combining just-in-time learning and one-class extreme learning | × | AUROC: 0.85 | |
| Our study | General ICU patients | 160,238 | Multicenter | pLOS-ICU | RF, SVM, DL, GBDT, and customized SAPS II | √ | - |
| Items | eICU-CRD | MIMIC-III |
|---|---|---|
| Country | United States | United States |
| Data | Multicenter | Single-center |
| Year | 2014–2015 | 2001–2012 |
| Number of units | 335 | 1 |
| Number of hospitals | 208 | 1 |
| Number of patients | 139,367 | 38,597 |
| Number of admissions | 200,859 | 53,423 |
| Deidentification | All protected health information was deidentified, and no patient privacy data can be identified. | |
| Data content | Vital sign measurements, laboratory tests, care plan documentation, diagnosis information, treatment information, and others. | |
| Items | eICU-CRD | MIMIC-III |
|---|---|---|
| Total number | 117,306 | 42,932 |
| Age/years | 61.6 ± 16.6 | 62.0 ± 16.5 |
| Gender, n (%) | ||
| Male | 64,244 (54.8%) | 24,740 (57.6%) |
| Female | 53,049 (45.2%) | 18,192 (42.4%) |
| SAPS II score | 30.0 ± 13.3 | 32.7 ± 12.7 |
| LOS-ICU (IQR1)/day | 1.8 (1.0–3.2) | 2.1 (1.2–4.0) |
| PLOS-ICU, n (%) | 31,296 (26.7%) | 14,951 (34.8%) |
| Models | eICU-CRD | MIMIC-III | ||||||
|---|---|---|---|---|---|---|---|---|
| Brier Score | AUROC | AUPRC | ECI | Brier Score | AUROC | AUPRC | ECI | |
| Customized SAPS II | 0.181 | 0.667 | 0.439 | 9.028 | 0.175 | 0.669 | 0.402 | 8.742 |
| RF | 0.166 | 0.735 | 0.530 | 8.317 | 0.169 | 0.745 | 0.530 | 8.469 |
| SVM | 0.183 | 0.690 | 0.480 | 9.137 | 0.172 | 0.716 | 0.482 | 8.577 |
| DL | 0.164 | 0.742 | 0.536 | 8.223 | 0.171 | 0.743 | 0.527 | 8.551 |
| GBDT | 0.164 | 0.742 | 0.537 | 8.224 | 0.166 | 0.747 | 0.536 | 8.294 |
| Ranks | GBTD | SAPS II |
|---|---|---|
| 1 | Pao2/Fio2 ratio | Glasgow Coma Score |
| 2 | Glasgow Coma Score | Age |
| 3 | Serum urea nitrogen level | Chronic diseases |
| 4 | Systolic blood pressure | Systolic blood pressure |
| 5 | White blood cell count | White blood cell count |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Lin, Y.; Li, P.; Hu, Y.; Zhang, L.; Kong, G. Predicting Prolonged Length of ICU Stay through Machine Learning. Diagnostics 2021, 11, 2242. https://doi.org/10.3390/diagnostics11122242
Wu J, Lin Y, Li P, Hu Y, Zhang L, Kong G. Predicting Prolonged Length of ICU Stay through Machine Learning. Diagnostics. 2021; 11(12):2242. https://doi.org/10.3390/diagnostics11122242
Chicago/Turabian StyleWu, Jingyi, Yu Lin, Pengfei Li, Yonghua Hu, Luxia Zhang, and Guilan Kong. 2021. "Predicting Prolonged Length of ICU Stay through Machine Learning" Diagnostics 11, no. 12: 2242. https://doi.org/10.3390/diagnostics11122242
APA StyleWu, J., Lin, Y., Li, P., Hu, Y., Zhang, L., & Kong, G. (2021). Predicting Prolonged Length of ICU Stay through Machine Learning. Diagnostics, 11(12), 2242. https://doi.org/10.3390/diagnostics11122242






