Cholera Rapid Diagnostic Tests for the Detection of Vibrio cholerae O1: An Updated Meta-Analysis
Abstract
:1. Introduction
2. Methods
2.1. Data Sources and Searches
2.2. Outcomes
2.3. Data Extraction and Quality Assessment
2.4. Data Analysis
3. Results
3.1. Literature Search
3.2. Characteristics of Included Studies
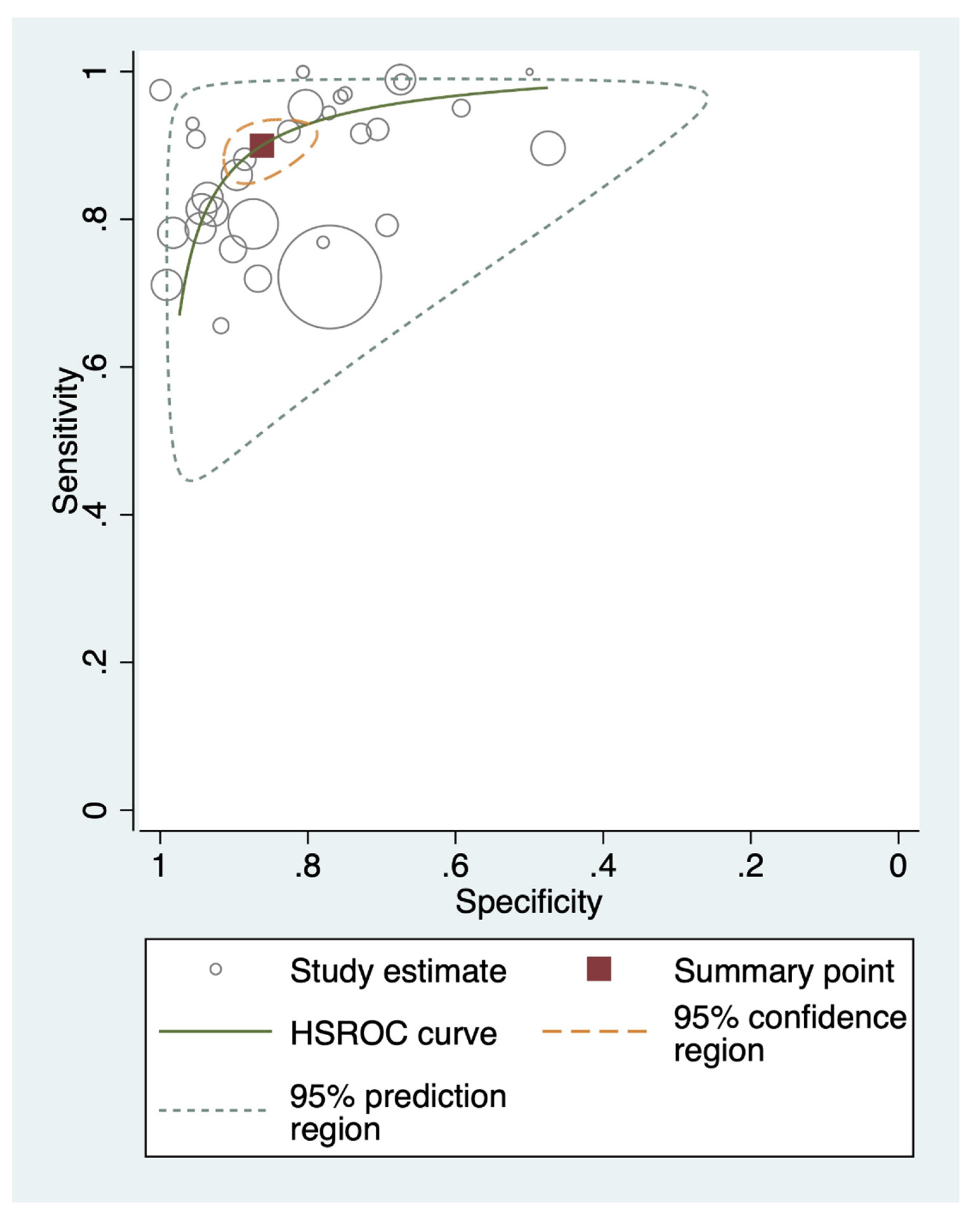
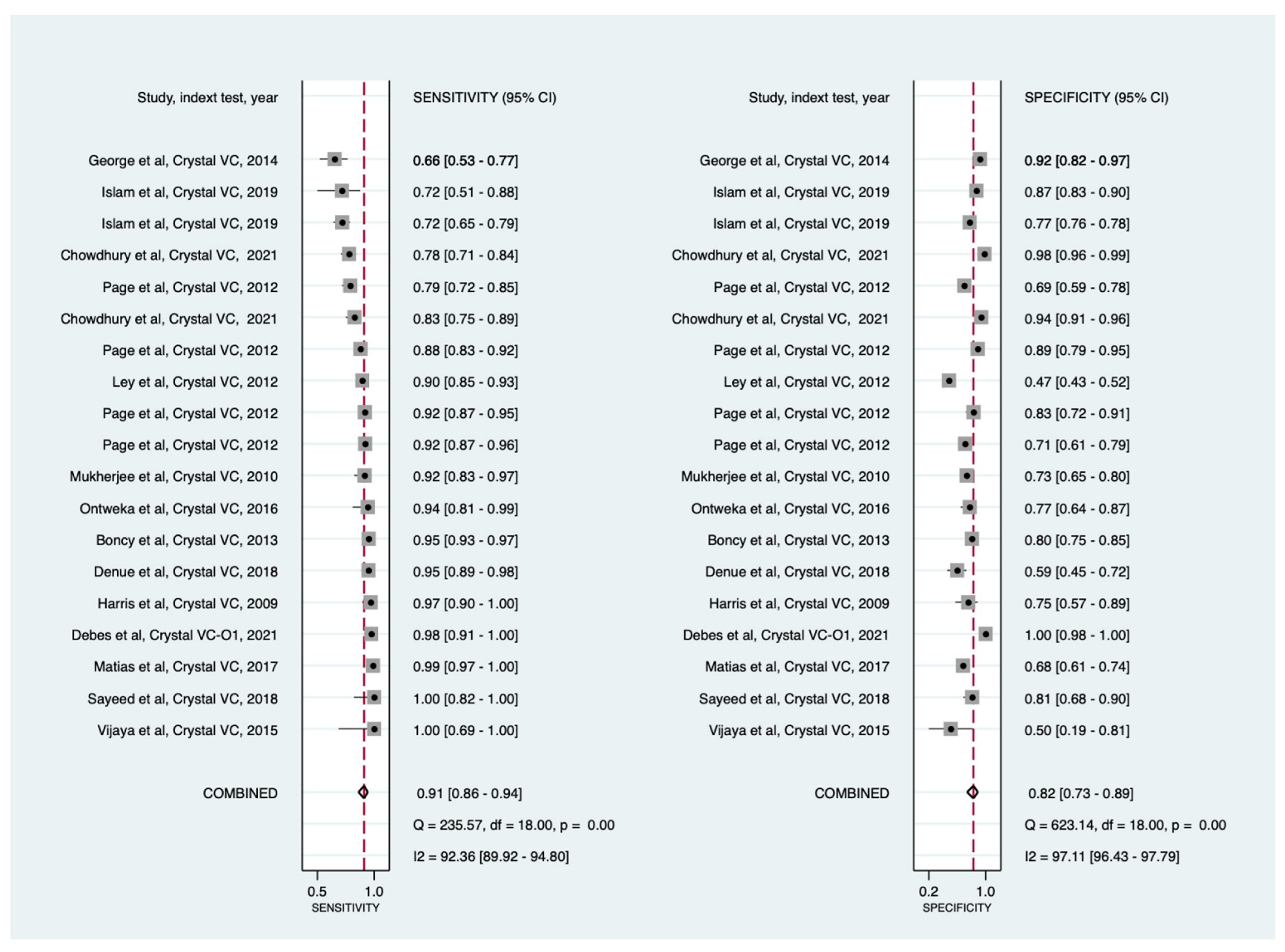
3.3. Meta-Analysis
3.3.1. Overall Performance
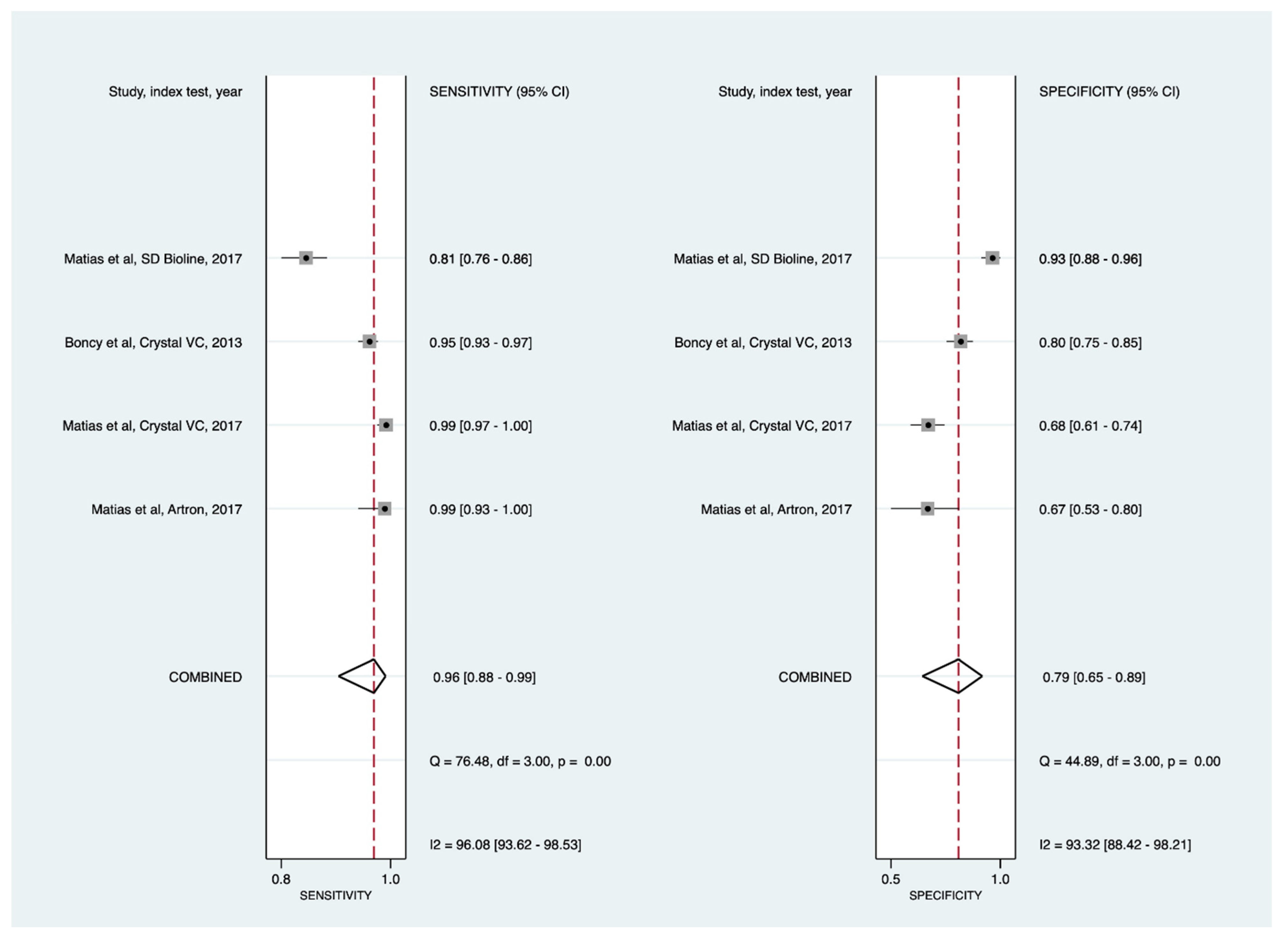
3.3.2. Sensitivity Analyses
Crystal VC RDTs
Cholera RDTs by Geographic Regions
Direct and APW Enrichment Testing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Study | Location | Study Period | Study Design | Participants’ Age (Year; Mean or Median)/Descriptor | User | Industry Funded (Yes or No) | Population | Specimen Type | Index Test | Reference Standard | Sample Size |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Debes et al., 2021 [13] | Kenya | 2018 to 2019 | Cross-sectional | All age groups (mean = 25) | Lab technician | No, but received RDT kit from the manufacturer | Hospital samples: Individuals presenting to a health facility with acute watery diarrhea. | Stool | Crystal VC-O1 | Culture and PCR | 230 |
| Chowdhury et al., 2021 [14] | India | 2016 and 2017 | Cross-sectional | All age groups | Lab technician | No | Hospital samples: Individuals hospitalized for diarrhea and children treated for diarrhea as outpatients at designated hospitals. | Stool | SD bioline cholera, SMART-II Cholera O1 and Crystal VC | Culture and PCR | 506 |
| Chibwe et al., 2020 [16] | Malawi | 2018 | Cross-sectional | All age groups (<5:14%; and >5: 86%) | Lab technician | No, but received RDT kit as a gift | Hospital samples: Individuals presenting to cholera treatment camps with acute diarrhea. | Stool (bulk stool or rectal swabs) | Cholkit | Culture | 80 |
| Islam et al., 2019 [17] | Bangladesh | Ongoing surveillance since 2016 | Cross-sectional | Mean = 19 | Lab technician | Yes | Hospital samples: Individuals presenting to hospitals with acute watery diarrhea. | Stool | Crystal VC | Culture | 381 |
| 5865 | |||||||||||
| 614 | |||||||||||
| Stool | Cholkit | Culture | 381 | ||||||||
| 1355 | |||||||||||
| 424 | |||||||||||
| Mwaba et al., 2018 [19] | Zambia | 2016 | Cross-sectional | Mean = 24 | Lab technician | Not reported | Hospital samples: Patients with acute non-blood watery diarrhea. | Stool | SD Bioline cholera | Culture | 170 |
| Denue, 2018 [21] | Nigeria | 2017 | Cross-sectional | Mean = 20 | Lab technician | No | Hospital samples: Individuals presenting to a cholera treatment unit with diarrhea. | Stool | Crystal VC | Culture | 156 |
| Sayeed et al., 2018 [18] | Bangladesh | Not reported | Cross-sectional | Median = 26 | Lab technician | Yes | Hospital samples: Patients presenting to the icddr, b hospital with acute watery diarrhea. | Stool | Cholkit | Culture | 76 |
| Stool | Crystal VC | 76 | |||||||||
| Matias et al., 2017 [31] | Haiti | 2014–2015 | Cross-sectional | Not reported | Lab technician | No | Hospital samples: Patients presenting to a cholera treatment center with acute watery diarrhea. | Stool | Crystal VC | Culture | 511 |
| Artron | Culture | 129 | |||||||||
| SD Bioline | Culture | 451 | |||||||||
| Bwire et al., 2017 [30] | Uganda | 2015 | Cross-sectional | All age groups | Lab technician | No | Hospital samples: Suspected cholera patients presenting to hospitals. | Stool/rectal swabs | Crystal VC | Culture | 102 |
| Ontweka et al., 2016 [32] | South Sudan | 2015 | Cross-sectional | Median = 26 | Lab technician | No | Hospital samples: Patients presenting to cholera treatment centers with acute watery diarrhea. | Stool | Crystal VC | PCR | 101 |
| Debes et al., 2016 [20] | Cameroon | 2013–2014 | Cross-sectional | All age groups | Lab technician | No | Hospital samples: Patients with acute non-blood watery diarrhea. | Stool | Crystal VC | PCR | 673 |
| Vijaya et al., 2015 [15] | India | Not reported | Cross-sectional | Not reported | Researcher | Not reported | Hospital samples: Patients presenting to a tertiary care hospital with acute watery diarrhea | Stool (18 bulk stool samples and 2 rectal swabs) | Crystal VC | Culture | 20 |
| George et al., 2014 [22] | Bangladesh | 2013 | Cross-sectional | Median = 32 | Lab technician | Not reported | Hospital samples: Patients presenting to the icddr, b hospital with moderate to severe dehydration and acute watery diarrhea. | Stool | Crystal VC | Culture | 125 |
| Boncy et al., 2013 [23] | Haiti | 2011 | Cross-sectional | Not reported | Lab technician | Not reported | Hospital samples: Patients with acute rice watery diarrhea. | Stool | Crystal VC | Culture | 644 |
| Ley et al., 2012 [24] | Tanzania | 2009 | Cross-sectional | Not reported | Lab technician/Field workers | No | Hospital samples: Patients presenting to treatment centers with watery diarrhea. | Stool | Crystal VC | Culture | 622 |
| Page et al., 2012 [25] | DR Congo | 2008 | Cross-sectional | >5 | Lab technician/Field worker | No | Hospital samples: Patients presenting to cholera treatment centers with acute watery diarrhea. | Stool | Crystal VC | Culture | 256 |
| Culture and/or PCR | 256 | ||||||||||
| Mukherjee et al., 2010 [26] | India | 2008 | Cross-sectional | All age groups | Lab technician | Not reported | Hospital samples: Hospitalized patients with diarrhea. | Stool | Crystal VC | Culture | 212 |
| Harris et al., 2009 [27] | Guinea-Bissau | 2008 | Cross-sectional | Median = 27 | Lab technician | Not reported | Hospital samples: Patients presenting to a hospital cholera ward. | Stool | Crystal VC | PCR | 101 |
| Wang et al., 2006 [28] | Mozambique | 2004 | Cross-sectional | Mean = 20 (cholera) and 24 (non-cholera) | Lab technician | No | Hospital samples: Patients with acute non-blood watery diarrhea. | Bulk stool | Institut Pasteur cholera dipstick | Culture | 172 |
| Rectal swabs | Institut Pasteur cholera dipstick | Culture | 219 | ||||||||
| Bhuiyan et al., 2003 [29] | Bangladesh | 2002 | Cross-sectional | 24 | Lab technician | Not reported | Hospital samples: Patients hospitalized at the icddr, b for diarrhea. | Rectal swabs | Institut Pasteur cholera dipstick | Culture | 134 |
| Study | Index Test | Sample Tested | Direct Specimen or after Enrichment | Results, n | |||
|---|---|---|---|---|---|---|---|
| True Positive | False Positive | False Negative | True Negative | ||||
| Debes et al., 2021 [13] | Crystal VC-O1 | Stool | Direct | 79 | 0 | 2 | 149 |
| Chowdhury et al., 2021 [14] | SD Bioline | Stool | Direct * | 115 | 11 | 105 | 275 |
| SMART-II | Stool | Direct * | 128 | 22 | 92 | 264 | |
| Crystal VC | Stool | Direct * | 122 | 9 | 98 | 277 | |
| Chowdhury et al., 2021 [14] | SD Bioline | Stool | Direct | 105 | 21 | 24 | 356 |
| Direct ** | 111 | 3 | 45 | 347 | |||
| SMART-II | Stool | Direct | 111 | 39 | 18 | 338 | |
| Direct ** | 123 | 19 | 33 | 331 | |||
| Crystal VC | Stool | Direct | 107 | 24 | 22 | 353 | |
| Direct ** | 122 | 6 | 34 | 344 | |||
| Chibwe et al., 2020 [16] | Cholkit | Stool | Direct | 53 | 1 | 4 | 22 |
| Enrichment | 56 | 0 | 1 | 23 | |||
| Islam et al., 2019 [17] | Crystal VC | Stool | Direct | 18 | 47 | 7 | 309 |
| Direct | 117 | 1308 | 45 | 4395 | |||
| Enrichment | 17 | 9 | 8 | 347 | |||
| Enrichment | 28 | 53 | 13 | 520 | |||
| Cholkit | Stool | Direct | 19 | 35 | 6 | 321 | |
| Direct | 27 | 166 | 7 | 1155 | |||
| Enrichment | 16 | 21 | 9 | 335 | |||
| Enrichment | 20 | 22 | 10 | 372 | |||
| Mwaba et al., 2018 [19] | SD Bioline | Stool | Direct | 60 | 5 | 6 | 99 |
| Enrichment | 63 | 0 | 3 | 104 | |||
| Denue, 2018 [21] | Crystal VC | Stool | Direct | 97 | 22 | 5 | 32 |
| Sayeed et al., 2018 [18] | Cholkit | Stool | Direct | 19 | 11 | 2 | 44 |
| Crystal VC | Stool | Direct | 19 | 11 | 2 | 44 | |
| Matias et al., 2017 [31] | Crystal VC | Stool | Direct | 282 | 65 | 3 | 135 |
| SD Bioline | Stool | Direct | 197 | 15 | 46 | 193 | |
| Artron | Stool | Direct | 73 | 17 | 1 | 35 | |
| Bwire et al., 2017 [30] | Crystal VC | Stool/Rectal swabs | Enrichment | 91 | 1 | 1 | 9 |
| Ontweka et al., 2016 [32] | Crystal VC | Stool | Direct | 34 | 13 | 2 | 44 |
| Enrichment | 31 | 0 | 5 | 64 | |||
| Debes et al., 2016 [20] | Crystal VC | Stool | Enrichment | 25 | 3 | 7 | 638 |
| Vijaya et al., 2015 [15] | Crystal VC | Stool | Direct ** | 10 | 5 | 0 | 5 |
| George et al., 2014 [22] | Crystal VC | Stool | Direct | 42 | 5 | 22 | 56 |
| Enrichment | 48 | 1 | 16 | 60 | |||
| Boncy et al., 2013 [23] | Crystal VC | Stool | Direct | 381 | 48 | 19 | 196 |
| Ley et al., 2012 [24] | Crystal VC | Stool | Direct | 182 | 220 | 21 | 199 |
| Page et al., 2012 [25] | Crystal VC | Stool | Direct | 142 | 30 | 12 | 72 |
| Direct | 122 | 31 | 32 | 70 | |||
| Direct | 164 | 8 | 22 | 62 | |||
| Direct | 171 | 12 | 15 | 57 | |||
| Mukherjee et al., 2010 [26] | Crystal VC | Stool | Direct | 66 | 38 | 6 | 102 |
| Harris et al., 2009 [27] | Crystal VC | Stool | Direct | 65 | 8 | 2 | 24 |
| Wang et al., 2006 [28] | Institut Pasteur cholera dipstick | Stool/Rectal swabs | Direct | 57 | 10 | 2 | 31 |
| Direct | 10 | 13 | 3 | 46 | |||
| Enrichment | 45 | 3 | 0 | 40 | |||
| Enrichment | 19 | 1 | 2 | 109 | |||
| Bhuiyan et al., 2003 [29] | Institut Pasteur cholera dipstick | Rectal swabs | Enrichment | 65 | 5 | 3 | 61 |
| Study | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| Debes et al., 2021 [13] |  |  |  |  |  |  |  |
| Chowdhury et al., 2021 [14] |  |  |  |  |  |  |  |
| Islam et al., 2019 [17] |  |  |  |  |  |  |  |
| Mwaba et al., 2018 [19] |  |  |  |  |  |  |  |
| Denue BA, 2018 [21] |  |  |  |  |  |  |  |
| Chibwe et al., 2020 [16] |  |  |  |  |  |  |  |
| Sayeed et al., 2018 [18] |  |  |  |  |  |  |  |
| Matias et al., 2017 [31] |  |  |  |  |  |  |  |
| Bwire et al., 2017 [30] |  |  |  |  |  |  |  |
| Ontweka et al., 2016 [32] |  |  |  |  |  |  |  |
| Debes et al., 2016 [20] |  |  |  |  |  |  |  |
| Vijaya et al., 2015 [15] |  |  |  |  |  |  |  |
| George et al., 2014 [22] |  |  |  |  |  |  |  |
| Boncy et al., 2013 |  |  |  |  |  |  |  |
| Ley et al., 2012 |  |  |  |  |  |  |  |
| Page et al., 2012 [25] |  |  |  |  |  |  |  |
| Mukherjee et al., 2010 [26] |  |  |  |  |  |  |  |
| Harris et al., 2009 [27] |  |  |  |  |  |  |  |
| Wang et al., 2006 [28] |  |  |  |  |  |  |  |
| Bhuiyan et al., 2003 [29] |  |  |  |  |  |  |  |
References
- Clemens, J.D.; Nair, G.B.; Ahmed, T.; Qadri, F.; Holmgren, J. Cholera. The Lancet Cholera. Lancet 2017, 390, 1539–1549. [Google Scholar] [CrossRef]
- Richterman, A.; Sainvilien, D.R.; Eberly, L.; Ivers, L.C. Individual and Household Risk Factors for Symptomatic Cholera Infection: A Systematic Review and Meta-analysis. J. Infect. Dis. 2018, 218, S154–S164. [Google Scholar] [CrossRef] [PubMed]
- Mutreja, A.; Kim, D.W.; Thomson, N.R.; Connor, T.R.; Lee, J.H.; Kariuki, S.; Croucher, N.J.; Choi, S.Y.; Harris, S.R.; Lebens, M.; et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 2011, 477, 462–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azman, A.S.; Luquero, F.J.; Ciglenecki, I.; Grais, R.F.; Sack, D.A.; Lessler, J. The Impact of a One-Dose versus Two-Dose Oral Cholera Vaccine Regimen in Outbreak Settings: A Modeling Study. PLoS Med. 2015, 12, e1001867. [Google Scholar] [CrossRef] [Green Version]
- Keddy, K.H.; Sooka, A.; Parsons, M.B.; Njanpop-Lafourcade, B.M.; Fitchet, K.; Smith, A.M. Diagnosis of Vibrio cholerae O1 infection in Africa. J. Infect. Dis. 2013, 208, S23–S31. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Global Task Force on Cholera Control Surveillance Laboratory Working Group. The Use of Cholera Rapid Diagnostic Tests. Available online: https://www.gtfcc.org/wp-content/uploads/2019/10/gtfcc-interim-use-of-cholera-rapid-diagnostic-tests.pdf (accessed on 9 September 2021).
- Muzembo, B.A.; Kitahara, K.; Debnath, A.; Okamoto, K.; Miyoshi, S. Accuracy of cholera rapid diagnostic tests: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2021, in press. [Google Scholar] [CrossRef]
- McInnes, M.D.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Schünemann, H.J.; Oxman, A.D.; Brozek, J.; Glasziou, P.; Jaeschke, R.; Vist, G.E.; Williams, J.W., Jr.; Kunz, R.; Craig, J.; Montori, V.M.; et al. Grading quality of evidence and strength of recommendations for diagnostic tests and strategies. BMJ 2008, 336, 1106–1110. [Google Scholar] [CrossRef] [Green Version]
- Leeflang, M.M.G.; Deeks, J.J.; Takwoingi, Y.; Macaskill, P. Cochrane diagnostic test accuracy reviews. Syst. Rev. 2013, 2, 82. [Google Scholar] [CrossRef] [Green Version]
- Leeflang, M.M.G. Systematic reviews and meta-analyses of diagnostic test accuracy. Clin. Microbiol. Infect. 2014, 20, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Debes, A.K.; Murt, K.N.; Waswa, E.; Githinji, G.; Umuro, M.; Mbogori, C.; Roskosky, M.; Ram, M.; Shaffer, A.; Sack, D.A.; et al. Laboratory and Field Evaluation of the Crystal VC-O1 Cholera Rapid Diagnostic Test. Am. J. Trop. Med. Hyg. 2021, 104, 2017–2023. [Google Scholar] [CrossRef]
- Chowdhury, G.; Senapati, T.; Das, B.; Kamath, A.; Pal, D.; Bose, P.; Deb, A.; Paul, S.; Mukhopadhyay, A.K.; Dutta, S.; et al. Laboratory evaluation of the rapid diagnostic tests for the detection of Vibrio cholerae O1 using diarrheal samples. PLoS Negl. Trop. Dis. 2021, 15, e0009521. [Google Scholar] [CrossRef]
- Vijaya, D.; TA, D.D. Rapid detection of vibrio cholerae O1 And O139 In stool samples by one-step immunochromatographic dip-stick test. Int. J. Biol. Med. Res. 2015, 6, 4990–4992. [Google Scholar]
- Chibwe, I.; Kasambara, W.; Kagoli, M.; Milala, H.; Gondwe, C.; Azman, A.S. Field Evaluation of Cholkit Rapid Diagnostic Test for Vibrio Cholerae O1 During a Cholera Outbreak in Malawi, 2018. Open Forum Infect. Dis. 2020, 7, ofaa493. [Google Scholar] [CrossRef]
- Islam, T.; Khan, A.I.; Sayeed, A.; Amin, J.; Islam, K.; Alam, N.; Sultana, N.; Jahan, N.; Rashid, M.; Khan, Z.H.; et al. Field evaluation of a locally produced rapid diagnostic test for early detection of cholera in Bangladesh. PLoS Negl. Trop. Dis. 2019, 13, e0007124. [Google Scholar] [CrossRef]
- Sayeed, A.; Islam, K.; Hossain, M.; Akter, N.J.; Alam, N.; Sultana, N.; Khanam, F.; Kelly, M.; Charles, R.C.; Kováč, P.; et al. Development of a new dipstick (Cholkit) for rapid detection of Vibrio cholerae O1 in acute watery diarrheal stools. PLoS Negl. Trop. Dis. 2018, 12, e0006286. [Google Scholar] [CrossRef] [Green Version]
- Mwaba, J.; Ferreras, E.; Chizema-Kawesa, E.; Mwimbe, D.; Tafirenyika, F.; Rauzier, J.; Blake, A.; Rakesh, A.; Poncin, M.; Stoitsova, S.; et al. Evaluation of the SD bioline cholera rapid diagnostic test during the 2016 cholera outbreak in Lusaka, Zambia. Trop. Med. Int. Health 2018, 23, 834–840. [Google Scholar] [CrossRef]
- Debes, A.K.; Ateudjieu, J.; Guenou, E.; Ebile, W.; Sonkoua, I.T.; Njimbia, A.C.; Steinwald, P.; Ram, M.; Sack, D.A. Clinical and Environmental Surveillance for Vibrio cholerae in Resource Constrained Areas: Application During a 1-Year Surveillance in the Far North Region of Cameroon. Am. J. Trop. Med. Hyg. 2016, 94, 537–543. [Google Scholar] [CrossRef]
- Denue, B.A. Evaluation of a rapid dipstick test (Crystal Vc®) for the diagnosis of cholera in Maiduguri, Northeastern Nigeria. Arch. Med. Health Sci. 2018, 6, 24–27. [Google Scholar] [CrossRef]
- George, C.M.; Rashid, M.-U.; Sack, D.A.; Sack, R.B.; Saif-Ur-Rahman, K.M.; Azman, A.; Monira, S.; Bhuyian, S.I.; Mahmud, M.T.; Mustafiz, M.; et al. Evaluation of enrichment method for the detection of Vibrio cholerae O1 using a rapid dipstick test in Bangladesh. Trop. Med. Int. Health 2014, 19, 301–307. [Google Scholar] [CrossRef]
- Boncy, J.; Rossignol, E.; Dahourou, G.; Hast, M.; Buteau, J.; Stanislas, M.; Moffett, D.; Bopp, C.; Balajee, S.A. Performance and utility of a rapid diagnostic test for cholera: Notes from Haiti. Diagn. Microbiol. Infect. Dis. 2013, 76, 521–523. [Google Scholar] [CrossRef]
- Ley, B.; Khatib, A.M.; Thriemer, K.; Von Seidlein, L.; Deen, J.; Mukhopadyay, A.; Chang, N.-Y.; Hashim, R.; Schmied, W.; Busch, C.J.L.; et al. Evaluation of a rapid dipstick (Crystal VC) for the diagnosis of cholera in Zanzibar and a comparison with previous studies. PLoS ONE 2012, 7, e36930. [Google Scholar] [CrossRef]
- Page, A.-L.; Alberti, K.P.; Mondonge, V.; Rauzier, J.; Quilici, M.-L.; Guerin, P. Evaluation of a rapid test for the diagnosis of cholera in the absence of a gold standard. PLoS ONE 2012, 7, e37360. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, P.; Ghosh, S.; Ramamurthy, T.; Bhattacharya, M.K.; Nandy, R.K.; Takeda, Y.; Nair, G.B.; Mukhopadhyay, A.K. Evaluation of a rapid immunochromatographic dipstick kit for diagnosis of cholera emphasizes its outbreak utility. Jpn. J. Infect. Dis. 2010, 63, 234–238. [Google Scholar]
- Harris, J.R.; Cavallaro, E.C.; De Nóbrega, A.A.; Dos, S.; Barrado, J.C.; Bopp, C.; Parsons, M.B.; Djalo, D.; Fonseca, F.G.D.S.; Ba, U.; et al. Field evaluation of crystal VC Rapid Dipstick test for cholera during a cholera outbreak in Guinea-Bissau. Trop. Med. Int. Health 2009, 14, 1117–1121. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Ansaruzzaman, M.; Vaz, R.; Mondlane, C.; Lucas, M.E.S.; Von Seidlein, L.; Deen, J.L.; Ampuero, S.; Puri, M.; Park, T.; et al. Field evaluation of a rapid immunochromatographic dipstick test for the diagnosis of cholera in a high-risk population. BMC Infect. Dis. 2006, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Bhuiyan, N.A.; Qadri, F.; Faruque, A.S.G.; A Malek, M.; Salam, M.A.; Nato, F.; Fournier, J.M.; Chanteau, S.; Sack, D.A.; Nair, G.B. Use of dipsticks for rapid diagnosis of cholera caused by Vibrio cholerae O1 and O139 from rectal swabs. J. Clin. Microbiol. 2003, 41, 3939–3941. [Google Scholar] [CrossRef] [Green Version]
- Bwire, G.; Orach, C.G.; Abdallah, D.; Debes, A.K.; Kagirita, A.; Ram, M.; Sack, D.A. Alkaline peptone water enrichment with a dipstick test to quickly detect and monitor cholera outbreaks. BMC Infect. Dis. 2017, 17, 726. [Google Scholar] [CrossRef] [Green Version]
- Matias, W.R.; Julceus, E.F.; Abelard, C.; Mayo-Smith, L.M.; Franke, M.; Harris, J.B.; Ivers, L.C. Laboratory evaluation of immunochromatographic rapid diagnostic tests for cholera in Haiti. PLoS ONE 2017, 12, e0186710. [Google Scholar] [CrossRef]
- Ontweka, L.N.; Deng, L.O.; Rauzier, J.; Debes, A.K.; Tadesse, F.; Parker, L.A.; Wamala, J.F.; Bior, B.K.; Lasuba, M.; But, A.B.; et al. Cholera Rapid Test with Enrichment Step Has Diagnostic Performance Equivalent to Culture. PLoS ONE 2016, 11, e0168257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngwa, M.C.; Wondimagegnehu, A.; Okudo, I.; Owili, C.; Ugochukwu, U.; Clement, P.; Devaux, I.; Pezzoli, L.; Ihekweazu, C.; Jimme, M.A.; et al. The multi-sectorial emergency response to a cholera outbreak in Internally Displaced Persons camps in Borno State, Nigeria, 2017. BMJ Glob. Health 2020, 5, e002000. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.A.; Carpenter, C.; Newman, T.B. Understanding the direction of bias in studies of diagnostic test accuracy. Acad. Emerg. Med. 2013, 20, 1194–1206. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.J.; Grembi, J.A.; Chao, D.L.; Andrews, J.R.; Alexandrova, L.; Rodriguez, P.H.; Ramachandran, V.V.; Sayeed, M.A.; Wamala, J.F.; Debes, A.K.; et al. Gold Standard Cholera Diagnostics Are Tarnished by Lytic Bacteriophage and Antibiotics. J. Clin. Microbiol. 2020, 58, e00412-20. [Google Scholar] [CrossRef]
- Alam, M.; Hasan, N.A.; Sultana, M.; Nair, G.B.; Sadique, A.; Faruque, A.S.G.; Endtz, H.P.; Sack, R.B.; Huq, A.; Colwell, R.R.; et al. Diagnostic limitations to accurate diagnosis of cholera. J. Clin. Microbiol. 2010, 48, 3918–3922. [Google Scholar] [CrossRef] [Green Version]
- Bouzid, D.; Zanella, M.C.; Kerneis, S.; Visseaux, B.; May, L.; Schrenzel, J.; Cattoir, V. Rapid diagnostic tests for infectious diseases in the emergency department. Clin. Microbiol. Infect. 2021, 27, 182–191. [Google Scholar] [CrossRef] [Green Version]



| Study Characteristic | Studies of Cholera Rapid Tests (N = 20), n (%) | |
|---|---|---|
| Study design | Cross-sectional | 20 (100) |
| Industry funded | Yes | 2 (10.0) |
| No | 11 (55.5) | |
| Not reported | 7 (35.5) | |
| Specimen type | Stool | 20 (100) |
| Testing type | Direct stool testing | 11 (55.0) |
| Stool enrichment with alkaline peptone water | 3 (15.0) | |
| Both | 6 (30.0) | |
| Commercial brand | Crystal VC | 15 (75.0) |
| Crystal VC-O1 | 1 (5.0) | |
| Cholkit | 3 (15.0) | |
| Pasteur Cholera Dipstick | 3 (15.0) | |
| SD Bioline | 3 (15.0) | |
| Cholera Smart O1 | 1 (5.0) | |
| Smart II Cholera O1 | 1 (5.0) | |
| Vibrio cholera strain detected | Vibrio cholera O1 | 20 (100) |
| Setting | Africa | 11 (55.5) |
| Asia | 7 (35.0) | |
| Americas | 2 (10.0) | |
| Test | Data Point (n) | Sample Size (n) | Pooled Sensitivity (95% CI), % | Pooled Specificity (95% CI), % | Positive LR (95% CI) | Negative LR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|---|
| All * | 45 | 19,280 | 90 (86 to 93) | 91 (87 to 94) | 10 (7 to 15) | 0.11 (0.08 to 0.15) | 89 (56 to 142) |
| Direct fresh stool | 32 | 15,877 | 90 (86 to 93) | 86 (81 to 90) | 7 (5 to 9) | 0.12 (0.09 to 0.16) | 56 (37 to 86) |
| Outcome | Number of Studies (Number of Specimens) | Study Design | Factors that May Lower Certainty of Evidence | Test Accuracy Certainty of Evidence | ||||
|---|---|---|---|---|---|---|---|---|
| Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | ||||
| True positives (patients correctly identified as having with cholera) | 20 (15,877) | Cross-sectional | Serious a | Not serious | Very serious b | Serious c | Likely d | Moderate ⊕⊕⊕ |
| False negative (patients incorrectly identified as not having cholera) | 20 (15,877) | Cross-sectional | Serious a | Not serious | Very serious b | Serious c | Likely d | Moderate ⊕⊕⊕  |
| True negatives (patients correctly identified as not having cholera) | 20 (15,877) | Cross-sectional | Serious a | Not serious | Very serious b | Serious c | Likely d | Moderate ⊕⊕⊕  |
| False positives (patients incorrectly identified as having cholera) | 20 (15,877) | Cross-sectional | Serious a | Not serious | Very serious b | Serious c | Likely d | Moderate ⊕⊕⊕  |
| Subgroup | Data Point (n) | Sample Size (n) | Pooled Sensitivity (95% CI), % | Pooled Specificity (95% CI), % | Positive LR (95% CI) | Negative LR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|---|
| Africa * | 13 | 2644 | 92 (89 to 94) | 83 (71 to 91) | 6 (3 to 10) | 0.09 (0.06 to 0.14) | 59 (24 to 145) |
| Asia (Bangladesh and India) ** | 15 | 11,527 | 82 (77 to 87) | 90 (84 to 94) | 8 (5 to 13) | 0.20 (0.15 to 0.26) | 42 (26 to 69) |
| Americas (Haiti) *** | 4 | 1706 | 96 (88 to 99) | 79 (65 to 89) | 5 (3 to 6) | 0.05 (0.02 to 0.13) | 99 (52 to 187) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muzembo, B.A.; Kitahara, K.; Ohno, A.; Debnath, A.; Okamoto, K.; Miyoshi, S.-I. Cholera Rapid Diagnostic Tests for the Detection of Vibrio cholerae O1: An Updated Meta-Analysis. Diagnostics 2021, 11, 2095. https://doi.org/10.3390/diagnostics11112095
Muzembo BA, Kitahara K, Ohno A, Debnath A, Okamoto K, Miyoshi S-I. Cholera Rapid Diagnostic Tests for the Detection of Vibrio cholerae O1: An Updated Meta-Analysis. Diagnostics. 2021; 11(11):2095. https://doi.org/10.3390/diagnostics11112095
Chicago/Turabian StyleMuzembo, Basilua Andre, Kei Kitahara, Ayumu Ohno, Anusuya Debnath, Keinosuke Okamoto, and Shin-Ichi Miyoshi. 2021. "Cholera Rapid Diagnostic Tests for the Detection of Vibrio cholerae O1: An Updated Meta-Analysis" Diagnostics 11, no. 11: 2095. https://doi.org/10.3390/diagnostics11112095
APA StyleMuzembo, B. A., Kitahara, K., Ohno, A., Debnath, A., Okamoto, K., & Miyoshi, S.-I. (2021). Cholera Rapid Diagnostic Tests for the Detection of Vibrio cholerae O1: An Updated Meta-Analysis. Diagnostics, 11(11), 2095. https://doi.org/10.3390/diagnostics11112095





