Intraoperative Resting-State Functional Connectivity Based on RGB Imaging
Abstract
:1. Introduction
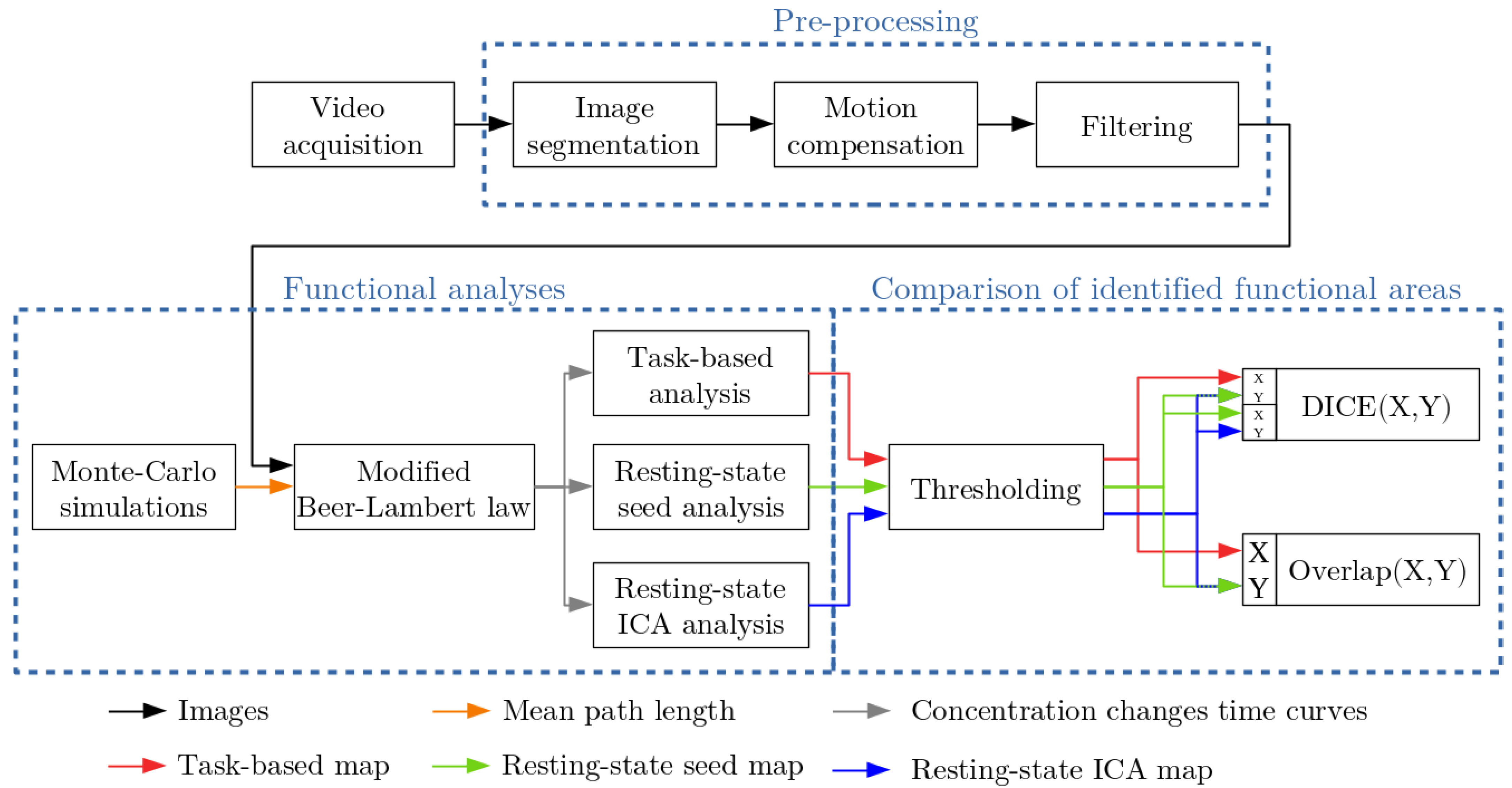
2. Material and Methods
2.1. Intraoperative Procedure
2.2. Functional Analyses
2.2.1. Task-Based Functional Analysis
2.2.2. Resting-State Analyses
2.2.3. Comparison of Identified Functional Areas
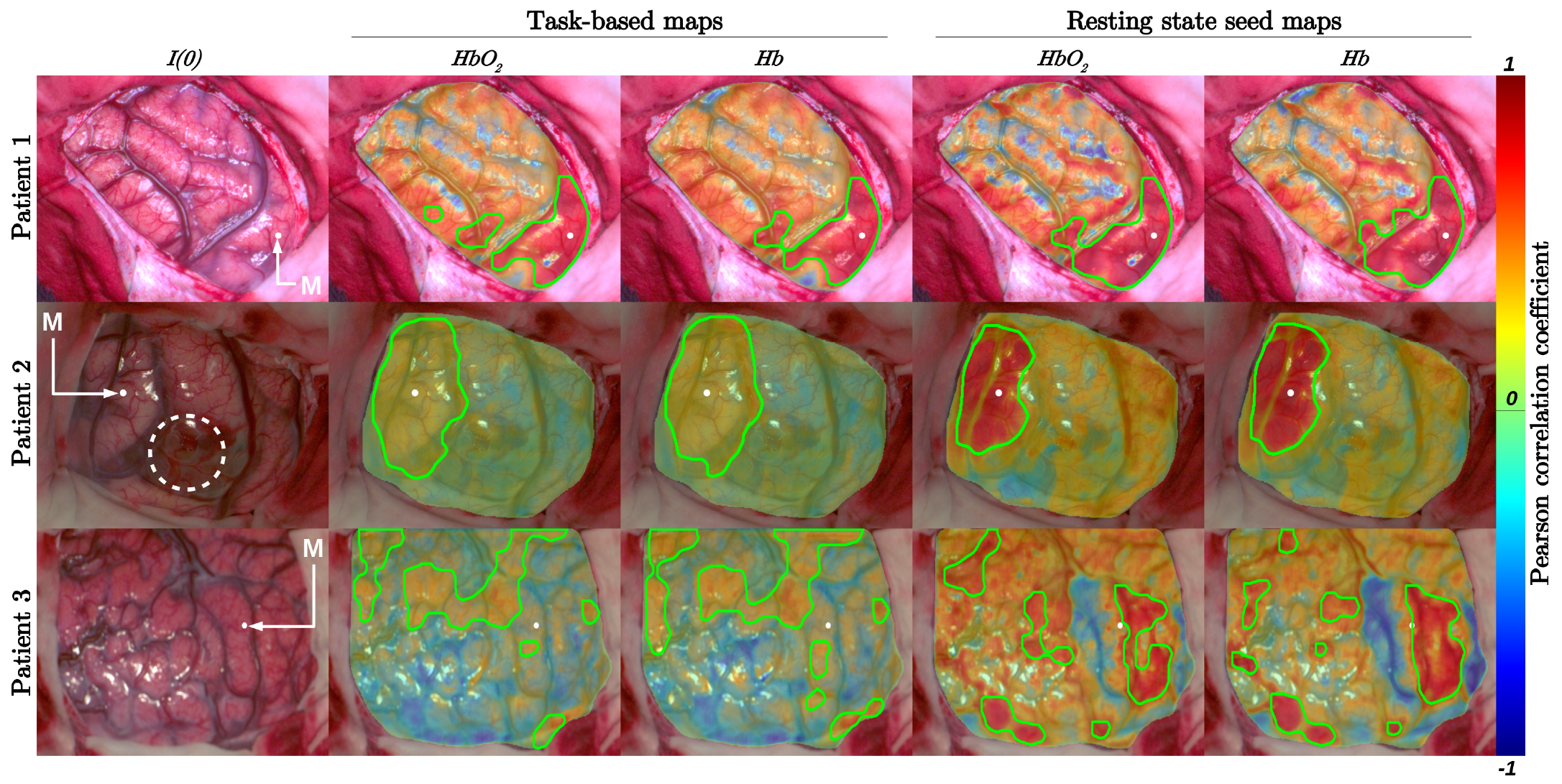
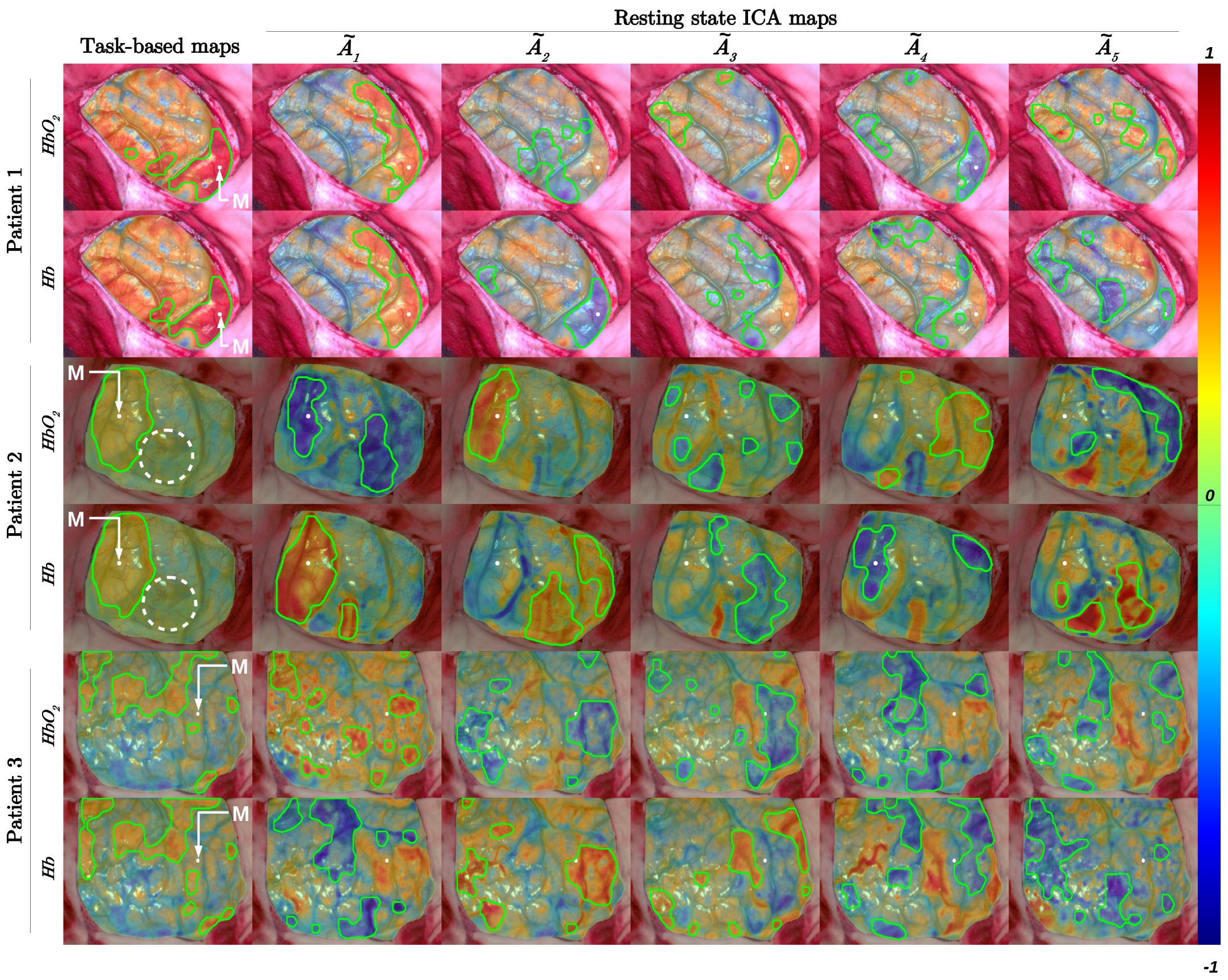
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| fMRI | functional Magnetic Resonance Imaging |
| fNIRS | functional Near Infra-Red Spectroscopy |
| EBS | Electrical Brain Stimulation |
| oxy hemoglobin | |
| deoxy hemoglobin | |
| tb | task-based |
| rs | resting-state |
| ICA | Independent Component Analysis |
References
- Ogawa, S.; Lee, T.M.; Kay, A.R.; Tank, D.W. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerard, I.J.; Kersten-Oertel, M.; Petrecca, K.; Sirhan, D.; Hall, J.A.; Collins, D.L. Brain shift in neuronavigation of brain tumors: A review. Med. Image Anal. 2017, 35, 403–420. [Google Scholar] [CrossRef]
- Penfield, W.; Boldrey, E. Somatic motor and sensory representation in the cerebral cortex of man as studied by electrical stimulation. Brain 1937, 60, 389–443. [Google Scholar] [CrossRef]
- Pallud, J.; Mandonnet, E.; Corns, R.; Dezamis, E.; Parraga, E.; Zanello, M.; Spena, G. Technical principles of direct bipolar electrostimulation for cortical and subcortical mapping in awake craniotomy. Neurochirurgie 2017, 63, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Zhuang, Z.; UnAh, C.; Alter, C.; Lipton, L. Cognition-activated low-frequency modulation of light absorption in human brain. Proc. Natl. Acad. Sci. USA 1993, 90, 3770–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oelschlägel, M.; Meyer, T.; Morgenstern, U.; Wahl, H.; Gerber, J.; Reiß, G.; Koch, E.; Steiner, G.; Kirsch, M.; Schackert, G.; et al. Mapping of language and motor function during awake neurosurgery with intraoperative optical imaging. Neurosurg. Focus FOC 2020, 48, E3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Alston, L.; Guyotat, J.; Montcel, B. Intraoperative quantitative functional brain mapping using an RGB camera. Neurophotonics 2019, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Guyotat, J.; Montcel, B. Real Time Intraoperative Functional Brain Mapping Based on RGB Imaging. IRBM 2021, 42, 189–197. [Google Scholar] [CrossRef]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Guyotat, J.; Montcel, B. Optimal Spectral Combination of a Hyperspectral Camera for Intraoperative Hemodynamic and Metabolic Brain Mapping. Appl. Sci. 2020, 10, 5158. [Google Scholar] [CrossRef]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Guyotat, J.; Montcel, B. Pixel-wise modified Beer-Lambert model for intraoperative functional brain mapping. In Clinical and Preclinical Optical Diagnostics II; Brown, J.Q., van Leeuwen, T.G., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2019; Volume 11073, pp. 148–152. [Google Scholar] [CrossRef]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Guyotat, J.; Montcel, B. Real time intraoperative functional brain mapping using a RGB camera. In Clinical and Preclinical Optical Diagnostics II; Brown, J.Q., van Leeuwen, T.G., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2019; Volume 11073, pp. 17–21. [Google Scholar] [CrossRef] [Green Version]
- Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Guyotat, J.; Montcel, B. Intraoperative functional and metabolic brain mapping using hyperspectral imaging. In Clinical and Translational Neurophotonics 2020; Madsen, S.J., Yang, V.X.D., Thakor, N.V., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2020; Volume 11225, pp. 24–30. [Google Scholar] [CrossRef]
- Fox, M.D.; Raichle, M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007, 8, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.; Zerrin Yetkin, F.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Pailler, M.; Faillenot, I.; Vassal, F.; Guyotat, J.; Barral, F.G.; Boutet, C. Presurgical Assessment of the Sensorimotor Cortex Using Resting-State fMRI. Am. J. Neuroradiol. 2016, 37, 101–107. [Google Scholar] [CrossRef] [Green Version]
- Signorelli, F.; Guyotat, J.; Schneider, F.; Isnard, J.; Bret, P. Technical Refinements for Validating Functional MRI-Based Neuronavigation Data by Electrical Stimulation during Cortical Language Mapping. Minim. Invasive Neurosurg. MIN 2003, 46, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Damoiseaux, J.S.; Rombouts, S.A.R.B.; Barkhof, F.; Scheltens, P.; Stam, C.J.; Smith, S.M.; Beckmann, C.F. Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. USA 2006, 103, 13848–13853. [Google Scholar] [CrossRef] [Green Version]
- Bisdas, S.; Charyasz-Leks, E.; Roder, C.; Tatagiba, M.S.; Ernemann, U.; Klose, U. Evidence of Resting-state Activity in Propofol-anesthetized Patients with Intracranial Tumors. Acad. Radiol. 2016, 23, 192–199. [Google Scholar] [CrossRef]
- Fukunaga, M.; Horovitz, S.G.; van Gelderen, P.; de Zwart, J.A.; Jansma, J.M.; Ikonomidou, V.N.; Chu, R.; Deckers, R.H.; Leopold, D.A.; Duyn, J.H. Large-amplitude, spatially correlated fluctuations in BOLD fMRI signals during extended rest and early sleep stages. Magn. Reson. Imaging 2006, 24, 979–992. [Google Scholar] [CrossRef]
- Mesquita, R.C.; Franceschini, M.A.; Boas, D.A. Resting state functional connectivity of the whole head with near-infrared spectroscopy. Biomed. Opt. Express 2010, 1, 324. [Google Scholar] [CrossRef] [PubMed]
- White, B.R.; Snyder, A.Z.; Cohen, A.L.; Petersen, S.E.; Raichle, M.E.; Schlaggar, B.L.; Culver, J.P. Resting-state functional connectivity in the human brain revealed with diffuse optical tomography. NeuroImage 2009, 47, 148–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Shaik, M.A.; Kozberg, M.G.; Kim, S.H.; Portes, J.P.; Timerman, D.; Hillman, E.M.C. Resting-state hemodynamics are spatiotemporally coupled to synchronized and symmetric neural activity in excitatory neurons. Proc. Natl. Acad. Sci. USA 2016, 113, E8463–E8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, M.K.; Kim, S.H.; Dovas, A.; Zhao, H.T.; Goldberg, A.R.; Xu, W.; Yagielski, A.J.; Cambareri, M.K.; Patel, K.B.; Mela, A.; et al. Glioma-Induced Alterations in Neuronal Activity and Neurovascular Coupling during Disease Progression. Cell Rep. 2020, 31, 107500. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dong, Q.; Niu, H. The minimum resting-state fNIRS imaging duration for accurate and stable mapping of brain connectivity network in children. Sci. Rep. 2017, 7, 6461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitlow, C.T.; Casanova, R.; Maldjian, J.A. Effect of Resting-State Functional MR Imaging Duration on Stability of Graph Theory Metrics of Brain Network Connectivity. Radiology 2011, 259, 516–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Q.; Boas, D.A. Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units. Opt. Express 2009, 17, 20178–20190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sdika, M.; Alston, L.; Rousseau, D.; Guyotat, J.; Mahieu-Williame, L.; Montcel, B. Repetitive motion compensation for real time intraoperative video processing. Med. Image Anal. 2019, 53, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sdika, M.; Alston, L.; Mahieu-Williame, L.; Guyotat, J.; Rousseau, D.; Montcel, B. Robust real time motion compensation for intraoperative video processing during neurosurgery. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1046–1049. [Google Scholar] [CrossRef]
- Oelschlägel, M.; Meyer, T.; Wahl, H.; Sobottka, S.B.; Kirsch, M.; Schackert, G.; Morgenstern, U. Evaluation of intraoperative optical imaging analysis methods by phantom and patient measurements. Biomed. Technol. Eng. 2013, 58, 257–267. [Google Scholar] [CrossRef]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 25, 120–123. [Google Scholar]
- Frigo, M.; Johnson, S.G. The Design and Implementation of FFTW3. Proc. IEEE 2005, 93, 216–231. [Google Scholar] [CrossRef] [Green Version]
- Veldsman, M.; Cumming, T.; Brodtmann, A. Beyond BOLD: Optimizing functional imaging in stroke populations: Optimizing BOLD Imaging in Stroke. Hum. Brain Mapp. 2015, 36, 1620–1636. [Google Scholar] [CrossRef] [PubMed]
- White, B.R.; Liao, S.M.; Ferradal, S.L.; Inder, T.E.; Culver, J.P. Bedside optical imaging of occipital resting-state functional connectivity in neonates. NeuroImage 2012, 59, 2529–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comon, P.; Jutten, C. Handbook of Blind Source Separation: Independent Component Analysis and Applications, 1st ed.; Academic Press, Inc.: Cambridge, MA, USA, 2010. [Google Scholar]
- Hyvarinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999, 10, 626–634. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Bruyns-Haylett, M.; Zheng, Y.; Berwick, J.; Jones, M. Temporal coupling between stimulus-evoked neural activity and hemodynamic responses from individual cortical columns. Phys. Med. Biol. 2010, 55, 2203–2219. [Google Scholar] [CrossRef]
- Stoecklein, V.M.; Stoecklein, S.; Galiè, F.; Ren, J.; Schmutzer, M.; Unterrainer, M.; Albert, N.L.; Kreth, F.W.; Thon, N.; Liebig, T.; et al. Resting-state fMRI detects alterations in whole brain connectivity related to tumor biology in glioma patients. Neuro-Oncol. 2020, 22, 1388–1398. [Google Scholar] [CrossRef]
- Friston, K.J. (Ed.) Statistical Parametric Mapping: The Analysis of Funtional Brain Images, 1st ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Boston, MA, USA, 2007. [Google Scholar]
- Li, Y.O.; Adali, T.; Calhoun, V.D. Estimating the number of independent components for functional magnetic resonance imaging data. Hum. Brain Mapp. 2007, 28, 1251–1266. [Google Scholar] [CrossRef]
| Patient 1 | Patient 2 | Patient 3 | ||
|---|---|---|---|---|
| Gender | Female | Male | Female | |
| Age | 37 | 57 | 45 | |
| Tumor | Low grade glioma | Lung cancer metastasis | Low grade glioma | |
| Surgical window | Right hemisphere | Right hemisphere | Right hemisphere | |
| General status | Awake | General anesthesia | Awake | |
| Task-based analysis | Task | Left-hand movement | Left-hand movement | Left-hand movement |
| performed by the patient | performed by an external person | performed by the patient | ||
| Number of cycles | 2 | 3 | 3 | |
| Acquisition duration | 1 min | 2 min | 2 min | |
| Resting-state analysis | Patient status | Looked at a medical practitioner | Under general anesthesia | Looked at a medical practitioner |
| and did not make any movements | and did not make any movements | and did not make any movements | ||
| Acquisition duration | 1 min 40 s | 2 min 20 s | 2 min 20 s | |
| Patient 1 | Patient 2 | Patient 3 | ||
|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.49 | 0.25 | 0.50 | 0.57 | 0.20 | 0.47 | 0.73 | 0.14 | 0.00 | 0.07 | 0.20 | 0.12 | 0.13 | 0.06 | 0.06 | ||
| 0.52 | 0.24 | 0.46 | 0.52 | 0.19 | 0.44 | 0.60 | 0.11 | 0.00 | 0.06 | 0.19 | 0.12 | 0.12 | 0.07 | 0.06 | ||
| 0.46 | 0.31 | 0.49 | 0.52 | 0.20 | 0.55 | 0.87 | 0.15 | 0.00 | 0.03 | 0.46 | 0.35 | 0.60 | 0.10 | 0.23 | ||
| 0.59 | 0.74 | 0.06 | 0.14 | 0.26 | 0.75 | 0.00 | 0.05 | 0.51 | 0.07 | 0.15 | 0.16 | 0.21 | 0.08 | 0.20 | ||
| 0.70 | 0.70 | 0.06 | 0.14 | 0.25 | 0.77 | 0.00 | 0.04 | 0.41 | 0.06 | 0.15 | 0.15 | 0.18 | 0.08 | 0.18 | ||
| 0.60 | 0.70 | 0.16 | 0.23 | 0.17 | 0.69 | 0.00 | 0.00 | 0.65 | 0.07 | 0.14 | 0.57 | 0.03 | 0.41 | 0.12 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caredda, C.; Mahieu-Williame, L.; Sablong, R.; Sdika, M.; Schneider, F.C.; Guyotat, J.; Montcel, B. Intraoperative Resting-State Functional Connectivity Based on RGB Imaging. Diagnostics 2021, 11, 2067. https://doi.org/10.3390/diagnostics11112067
Caredda C, Mahieu-Williame L, Sablong R, Sdika M, Schneider FC, Guyotat J, Montcel B. Intraoperative Resting-State Functional Connectivity Based on RGB Imaging. Diagnostics. 2021; 11(11):2067. https://doi.org/10.3390/diagnostics11112067
Chicago/Turabian StyleCaredda, Charly, Laurent Mahieu-Williame, Raphaël Sablong, Michaël Sdika, Fabien C. Schneider, Jacques Guyotat, and Bruno Montcel. 2021. "Intraoperative Resting-State Functional Connectivity Based on RGB Imaging" Diagnostics 11, no. 11: 2067. https://doi.org/10.3390/diagnostics11112067






