Increased Expression of Pyroptosis in Leukocytes of Patients with Kawasaki Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Experiment Design
2.2.1. DNA Methylation Profiling with Illumina M450K BeadChip
2.2.2. Gene Expression Profiling with Microarray
2.2.3. RNA Isolation and Real-Time Quantitative RT-PCR
2.2.4. Functional Study to Validate the Expression of CASP 1/4/5 in KD Patients
2.3. Statistical Analysis
3. Results
3.1. Promoter Hypomethylation and Upregulation of Caspases mRNA Levels in KD Patients
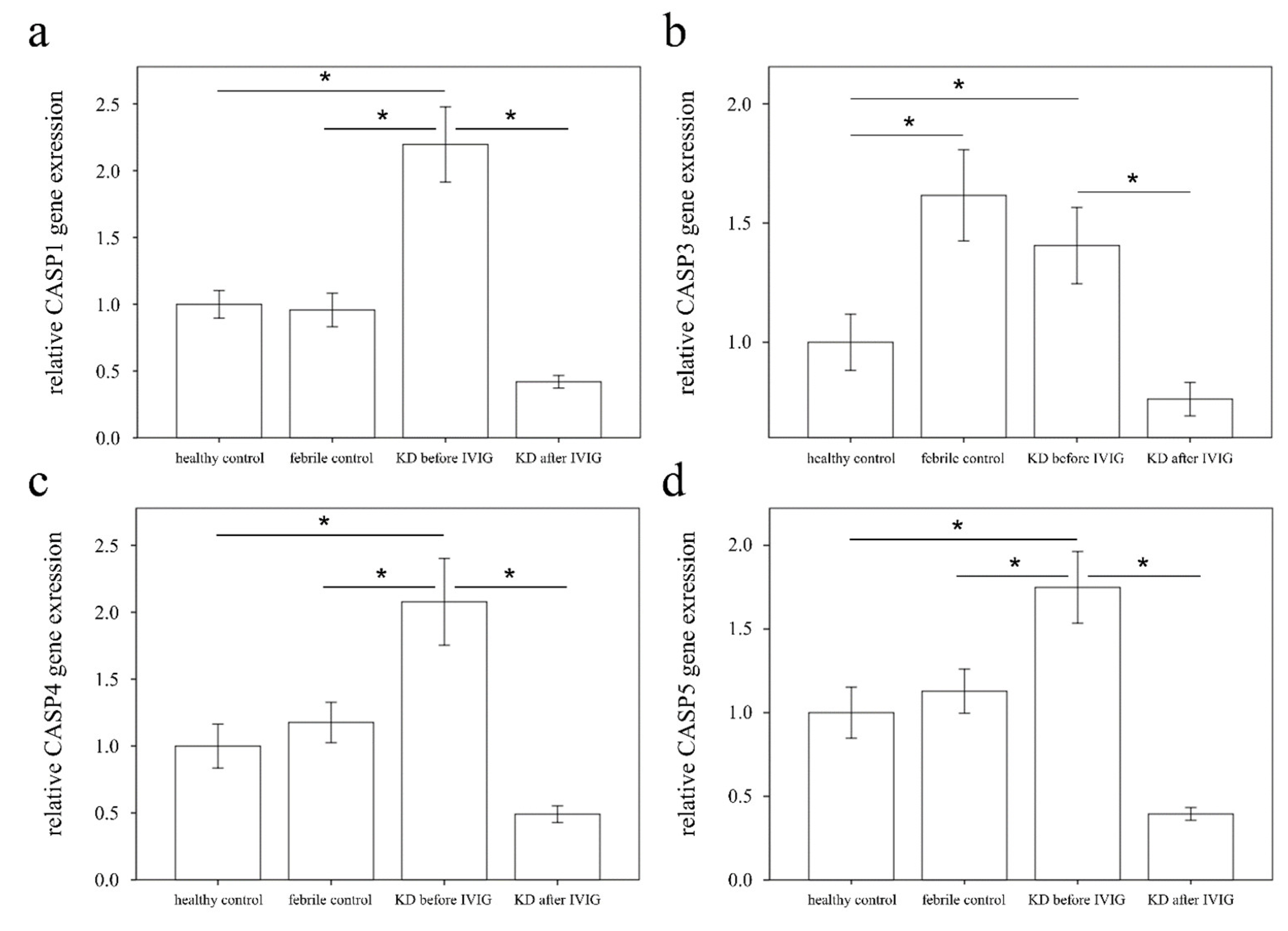
3.2. Increased CASP1, CASP3, CASP4, and CASP5 Expressions in the WBC of KD Patients
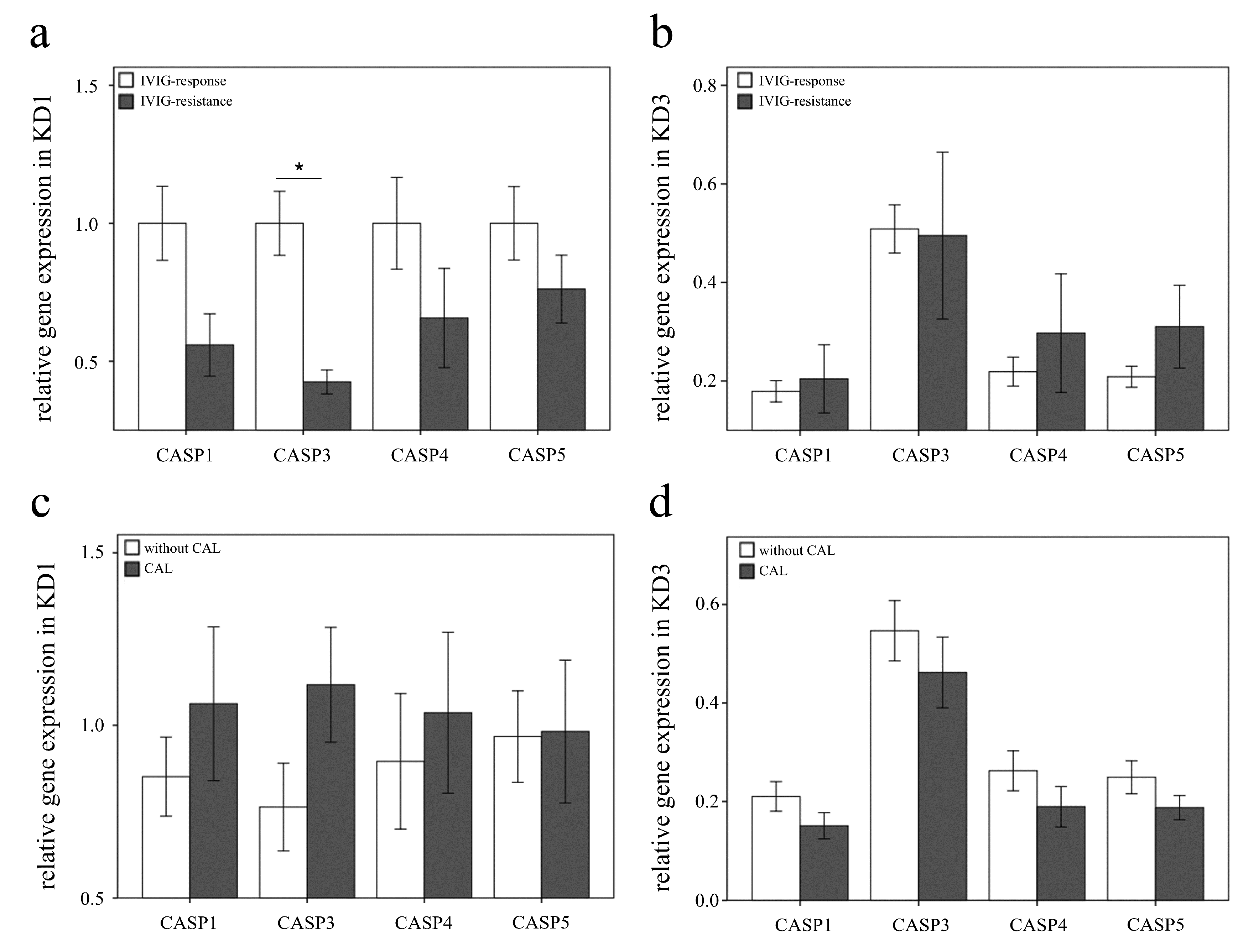
3.3. The Increased Expression of CASP5 in U937 Cells Stimulated with Plasma of KD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Y.-H.; Lin, K.-M.; Ho, S.-C.; Yan, J.-H.; Lo, M.-H.; Kuo, H.-C. Increased Incidence of Kawasaki Disease in Taiwan in Recent Years: A 15 Years Nationwide Population-Based Cohort Study. Front. Pediatr. 2019, 7, 121. [Google Scholar] [CrossRef]
- Wang, C.L.; Wu, Y.T.; Liu, C.A.; Kuo, H.C.; Yang, K.D. Kawasaki disease: Infection, immunity and genetics. Pediatr. Infect. Dis. J. 2005, 24, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Newburger, J.W.; Takahashi, M.; Beiser, A.S.; Burns, J.C.; Bastian, J.; Chung, K.J.; Rosen, F.S. A single intravenous infusion of gamma globulin as compared with four in-fusions in the treatment of acute Kawasaki syndrome. N. Engl. J. Med. 1991, 324, 1633–1639. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.-C.; Guo, M.M.-H.; Lo, M.-H.; Hsieh, K.-S.; Huang, Y.-H. Effectiveness of intravenous immunoglobulin alone and intravenous immunoglobulin combined with high-dose aspirin in the acute stage of Kawasaki disease: Study protocol for a randomized controlled trial. BMC Pediatr. 2018, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, R.; Hamada, H.; Sato, Y.; Suzuki, H.; Onouchi, Y.; Ebata, R.; Nagashima, K.; Terauchi, M.; Terai, M.; Hanaoka, H.; et al. Study protocol for a phase III multicentre, randomised, open-label, blinded-end point trial to evaluate the efficacy and safety of immunoglobulin plus cyclosporin A in patients with severe Kawasaki disease (KAICA Trial). BMJ Open 2015, 5, e009562. [Google Scholar] [CrossRef] [PubMed]
- Miura, M. Role of glucocorticoids in Kawasaki disease. Int. J. Rheum. Dis. 2017, 21, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Chen, K.-D.; Lo, M.-H.; Cai, X.-Y.; Kuo, H.-C. Decreased Steroid Hormone Receptor NR4A2 Expression in Kawasaki Disease Before IVIG Treatment. Front. Pediatr. 2019, 7, 7. [Google Scholar] [CrossRef]
- Jia, C.; Zhang, J.; Chen, H.; Zhuge, Y.; Chen, H.; Qian, F.; Zhou, K.; Niu, C.; Wang, F.; Qiu, H.; et al. Endothelial cell pyroptosis plays an important role in Kawasaki disease via HMGB1/RAGE/cathespin B signaling pathway and NLRP3 inflammasome activation. Cell Death Dis. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Gorelik, M.; Lee, Y.; Abe, M.; Andrews, T.; Davis, L.; Patterson, J.; Chen, S.; Crother, T.R.; Aune, G.; Rivas, M.N.; et al. IL-1 receptor antagonist, anakinra, prevents myocardial dysfunction in a mouse model of Kawasaki disease vasculitis and myocarditis. Clin. Exp. Immunol. 2019, 198, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, B.-C.; Gong, S.-J.; Yan, J. Homocysteine-Induced Caspase-3 Activation by Endoplasmic Reticulum Stress in Endothelial Progenitor Cells from Patients with Coronary Heart Disease and Healthy Donors. Biosci. Biotechnol. Biochem. 2011, 75, 1300–1305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wakita, D.; Kurashima, Y.; Crother, T.R.; Rivas, M.N.; Lee, Y.; Chen, S.; Fury, W.; Bai, Y.; Wagner, S.; Li, D.; et al. Role of Interleukin-1 Signaling in a Mouse Model of Kawasaki Disease–Associated Abdominal Aortic Aneurysm. Arter. Thromb. Vasc. Biol. 2016, 36, 886–897. [Google Scholar] [CrossRef]
- Koné-Paut, I.; Tellier, S.; Belot, A.; Brochard, K.; Guitton, C.; Marie, I.; Piedvache, C. Open Label, Phase II Study with Anakinra in Intravenous Immunoglobulin-Resistant Kawasaki Disease. Arthritis Rheumatol. 2021, 73, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Li, S.-C.; Huang, L.-H.; Chen, P.-C.; Lin, Y.-Y.; Lin, C.-C.; Kuo, H.-C. Identifying genetic hypomethylation and upregulation of toll-like receptors in Kawasaki disease. Oncotarget 2017, 8, 11249–11258. [Google Scholar] [CrossRef]
- Kuo, H.-C.; Li, S.-C.; Huang, L.-H.; Huang, Y.-H. Epigenetic hypomethylation and upregulation of matrix metalloproteinase 9 in Kawasaki disease. Oncotarget 2017, 8, 60875–60891. [Google Scholar] [CrossRef][Green Version]
- Huang, Y.-H.; Lo, M.-H.; Cai, X.-Y.; Kuo, H.-C. Epigenetic hypomethylation and upregulation of NLRC4 and NLRP12 in Kawasaki disease. Oncotarget 2018, 9, 18939–18948. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Lo, M.-H.; Cai, X.-Y.; Liu, S.-F.; Kuo, H.-C. Increase expression of CD177 in Kawasaki disease. Pediatr. Rheumatol. 2019, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Chen, K.-D.; Lo, M.-H.; Cai, X.-Y.; Chang, L.-S.; Kuo, Y.-H.; Huang, W.-D.; Kuo, H.-C. Decreased DNA methyltransferases expression is associated with coronary artery lesion formation in Kawasaki disease. Int. J. Med. Sci. 2019, 16, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.; Huang, Y.; Lin, T.; Du, Y.; Tsai, P.; Hsieh, C.; Chuang, J. Selective activation of Toll-like receptor 7 in activated hepatic stellate cells may modulate their profibrogenic phenotype. Biochem. J. 2012, 447, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Callaway, J.B.; Ting, J.P.-Y. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-Y.; Xie, K.-X.; Wang, S.-L.; Yuan, L.-W. Inflammatory caspase-related pyroptosis: Mechanism, regulation and therapeutic potential for inflammatory bowel disease. Gastroenterol. Rep. 2018, 6, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Velloso, F.J.; Lima, M.T.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39, BSR20181709. [Google Scholar] [CrossRef] [PubMed]
- Broz, P. Immunology: Caspase target drives pyroptosis. Nature 2015, 526, 642–643. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Z.; Yung, S.; Lu, Q. Epigenetic dynamics in immunity and autoimmunity. Int. J. Bio-Chem. Cell Biol. 2015, 67, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Hu, Q.; Long, H.; Chang, C.; Lu, Q.; Xiao, R. Epigenetic Variability of CD4+CD25+ Tregs Contributes to the Pathogenesis of Autoimmune Diseases. Clin. Rev. Allergy Immunol. 2016, 52, 260–272. [Google Scholar] [CrossRef]
- Li, S.-C.; Chan, W.-C.; Huang, Y.-H.; Guo, M.M.-H.; Yu, H.-R.; Huang, F.-C.; Kuo, H.-C.; Kuo, H.-C. Major methylation alterations on the CpG markers of inflammatory immune associated genes after IVIG treatment in Kawasaki disease. BMC Med. Genom. 2016, 9, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Hsu, Y.W.; Wu, C.M.; Chen, S.H.Y.; Hung, K.S.; Chang, W.P.; Chang, W.C. A replication study for association of ITPKC and CASP3 two-locus analysis in IVIG unresponsiveness and coronary artery lesion in Kawasaki disease. PLoS ONE 2013, 8, e69685. [Google Scholar] [CrossRef]
- Kuo, H.-C.; Yu, H.-R.; Juo, S.-H.H.; Yang, K.D.; Wang, Y.-S.; Liang, C.-D.; Chen, W.-C.; Chang, W.-P.; Huang, C.-F.; Lee, C.-P.; et al. CASP3 gene single-nucleotide polymorphism (rs72689236) and Kawasaki disease in Taiwanese children. J. Hum. Genet. 2010, 56, 161–165. [Google Scholar] [CrossRef]
- Onouchi, Y.; Suzuki, Y.; Suzuki, H.; Terai, M.; Yasukawa, K.; Hamada, H.; Suenaga, T.; Honda, T.; Honda, A.; Kobayashi, H.; et al. ITPKC and CASP3 polymorphisms and risks for IVIG unresponsiveness and coronary artery lesion formation in Kawasaki disease. Pharm. J. 2011, 13, 52–59. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Lin, M.-T.; Wang, J.-K.; Wu, M.-H. State-of-the-art acute phase management of Kawasaki disease after 2017 scientific statement from the American Heart Association. Pediatr. Neonatol. 2018, 59, 543–552. [Google Scholar] [CrossRef] [PubMed]
- McCrindle, B.W.; Rowley, A.; Newburger, J.W.; Burns, J.C.; Bolger, A.F.; Gewitz, M.; Baker, A.L.; Jackson, M.A.; Takahashi, M.; Shah, P.B.; et al. Diagnosis, Treatment, and Long-Term Management of Kawasaki Disease: A Scientific Statement for Health Professionals from the American Heart Association. Circulation 2017, 135, e927–e999. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-H.; Kuo, H.-C.; Pan, C.-T.; Lin, Y.-S.; Huang, Y.-H.; Li, S.-C. Multiomics analyses identified epigenetic modulation of the S100A gene family in Kawasaki disease and their significant involvement in neutrophil transendothelial migration. Clin. Epigenetics 2018, 10, 1–13. [Google Scholar] [CrossRef]
- Li, C.; Yin, W.; Yu, N.; Zhang, D.; Zhao, H.; Liu, J.; Lin, L. miR-155 promotes macrophage pyroptosis induced by Porphyromonas gingivalis through regulating the NLRP3 inflammasome. Oral Dis. 2019, 25, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, D.M.; Pereira, M.S.F.; Silva, A.L.N.; Cunha, L.D.; Zamboni, D.S. Caspase-1 but Not Caspase-11 Is Required for NLRC4-Mediated Pyroptosis and Restriction of Infection by Flagellated Legionella Species in Mouse Macrophages and In Vivo. J. Immunol. 2015, 195, 2303–2311. [Google Scholar] [CrossRef]
- Lacey, C.A.; Mitchell, W.J.; Dadelahi, A.S.; Skyberg, J.A. Caspases-1 and caspase-11 mediate pyroptosis, inflammation, and control of Brucella joint infection. Infect. Immun. 2018, 86, e00361-18. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.T.; Sze, D.M.-Y.; Chan, K.H.; Leung, P.H.-M. Involvement of caspase-4 in IL-1 beta production and pyroptosis in human macrophages during dengue virus infection. Immunobiology 2017, 223, 356–364. [Google Scholar] [CrossRef]
- Sarhan, J.; Liu, B.C.; Muendlein, H.I.; Li, P.; Nilson, R.; Tang, A.Y.; Poltorak, A. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. USA 2018, 115, E10888–E10897. [Google Scholar] [CrossRef]
- Chen, D.; Texada, D.E.; Duggan, C.; Deng, Y.; Redens, T.B.; Langford, M.P. Caspase-3 and -7 mediate apoptosis of human Chang’s conjunctival cells induced by enterovirus 70. Virology 2006, 347, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Yamamura, K.; Sakai, Y. The up-to-date pathophysiology of Kawasaki disease. Clin. Transl. Immunol. 2021, 10, e1284. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Pi, S.; Nie, G.; Wei, Z.; Yang, F.; Wang, C.; Xing, C.; Hu, G.; Zhang, C. Inhibition of ROS/NLRP3/Caspase-1 mediated pyroptosis alleviates excess molybdenum-induced apoptosis in duck renal tubular epithelial cells. Ecotoxicol. Environ. Saf. 2020, 208, 111528. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Yang, Y.-L.; Huang, F.-C.; Tiao, M.-M.; Lin, Y.-C.; Tsai, M.-H.; Wang, F.-S. MicroRNA-29a mitigation of endoplasmic reticulum and autophagy aberrance counteracts in obstructive jaundice-induced fibrosis in mice. Exp. Biol. Med. 2017, 243, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; He, M.; Shimizu, C.; Croker, B.A.; Hoffman, H.M.; Tremoulet, A.H.; Shyy, J.Y.J. Inflammasome Activation in Children with Kawasaki Disease and Multisystem Inflammatory Syndrome. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2509–2511. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.J.; Boucher, D.; Bierschenk, D.; Tebartz, C.; Whitney, P.; D’Silva, D.B.; Tanzer, M.C.; Monteleone, M.; Robertson, A.; Cooper, M.; et al. NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur. J. Immunol. 2015, 45, 2918–2926. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, Y.; Gao, W.; Ding, J.; Li, P.; Hu, L.; Shao, F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014, 514, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Sagulenko, V.; Vitak, N.; Vajjhala, P.R.; Vince, J.E.; Stacey, K.J. Caspase-1 Is an Apical Caspase Leading to Caspase-3 Cleavage in the AIM2 Inflammasome Response, Independent of Caspase-8. J. Mol. Biol. 2018, 430, 238–247. [Google Scholar] [CrossRef]
- Onouchi, Y. The genetics of Kawasaki disease. Int. J. Rheum. Dis. 2017, 21, 26–30. [Google Scholar] [CrossRef]
- Balsam, L.B.; Mokhtari, G.K.; Jones, S.; Peterson, S.; Hoyt, E.G.; Kofidis, T.; Robbins, R.C. Early inhibition of caspase-3 activity lessens the development of graft coronary artery disease. J. Heart Lung Transplant. 2005, 24, 827–832. [Google Scholar] [CrossRef] [PubMed]



| Gene Symbol | Accession Number | Hybridization | Primers (5′ to 3′) |
|---|---|---|---|
| RNA18S5 | NR_003286.2 | forward | GTAACCCGTTGAACCCCATT |
| reverse | CCATCCAATCGGTAGTAGCG | ||
| CASP1 | NM_001223 | forward reverse | TTTCCGCAAGGTTCGATTTTCA GGCATCTGCGCTCTACCATC |
| CASP3 | NM_004346 | forward | AGCGAATCAATGGACTCTGGA |
| reverse | GGTTTGCTGCATCGACATCT | ||
| CASP4 | NM_001225 | forward | CAAGAGAAGCAACGTATGGCA |
| reverse | AGGCAGATGGTCAAACTCTGTA | ||
| CASP5 | NM_001136112 | forward | ATGGCATCCTAGAGGGAATCT |
| reverse | ATGGCATCCTAGAGGGAATCT |
| Symbol | RefSeq | Column ID | Fold-Change (KD1 vs. HC) | p Value (KD1 vs. HC) | Fold-Change (KD1 vs. FC) | p Value (KD1 vs. FC) | Fold-Change (KD3 vs. KD1) | p Value (KD3 vs. KD1) |
|---|---|---|---|---|---|---|---|---|
| CASP1 | NM_001223 | TC11003505.hg.1 | 1.993 | 0.011 * | 1.445 | 0.118 | −2.443 | 0.003 * |
| CASP2 | NM_032982 | TC07000932.hg.1 | 1.172 | 0.058 | −1.026 | 0.725 | −1.303 | 0.006 * |
| CASP3 | NM_004346 | TC04001807.hg.1 | 1.471 | 0.035 * | 1.182 | 0.306 | −1.495 | 0.030 * |
| CASP4 | NM_001225 | TC11002246.hg.1 | 1.785 | 0.019 * | 1.346 | 0.169 | −2.235 | 0.003 * |
| CASP5 | NM_001136112 | TC11002247.hg.1 | 3.417 | 0.001 * | 2.053 | 0.022 * | −3.811 | 0.001 * |
| CASP6 | NM_001226 | TC04001464.hg.1 | 1.076 | 0.119 | −1.104 | 0.046 * | −1.175 | 0.005 * |
| CASP7 | NM_001227 | TC10000825.hg.1 | −1.002 | 0.890 | −1.039 | 0.062 | −1.047 | 0.030 * |
| CASP8 | NM_001080124 | TC02001177.hg.1 | 1.091 | 0.371 | 1.047 | 0.630 | −1.039 | 0.685 |
| CASP8AP2 | NM_001137667 | TC06000800.hg.1 | −1.102 | 0.565 | 1.002 | 0.990 | −1.069 | 0.691 |
| CASP9 | NM_001229 | TC01002245.hg.1 | 1.014 | 0.794 | −1.055 | 0.321 | −1.039 | 0.469 |
| CASP10 | NM_032976 | TC02001176.hg.1 | 1.369 | 0.032 * | −1.010 | 0.935 | −1.535 | 0.008 * |
| CASP12 | NM_001191016 | TC11002244.hg.1 | 1.016 | 0.796 | −1.058 | 0.366 | −1.005 | 0.935 |
| CASP14 | NM_012114 | TC19000266.hg.1 | −1.058 | 0.619 | −1.146 | 0.250 | 1.066 | 0.577 |
| CASP16 | ENST00000428155 | TC16000103.hg.1 | −1.033 | 0.792 | −1.070 | 0.585 | 1.118 | 0.378 |
| PYCARD | NM_013258 | TC16001052.hg.1 | 1.183 | 0.160 | −1.033 | 0.774 | −1.174 | 0.179 |
| IL-1α | NM_000575 | TC02002218.hg.1 | −1.080 | 0.335 | 1.030 | 0.706 | 1.067 | 0.411 |
| GSDMD | NM_001166237 | TC08000824.hg.1 | 1.101 | 0.153 | −1.022 | 0.726 | −1.086 | 0.214 |
| NLRP3 | NM_001079821 | TC01002008.hg.1 | −1.040 | 0.750 | 1.131 | 0.325 | −1.046 | 0.715 |
| IL-1β | NM_000576 | TC02002219.hg.1 | 1.677 | 0.066 | 2.195 | 0.012 * | −2.475 | 0.006 * |
| IL-18 | NM_001562 | TC11002293.hg.1 | 1.411 | 0.012 * | 1.115 | 0.338 | −1.417 | 0.011 * |
| Characteristic | Healthy Controls (n = 44) | Febrile Controls (n = 44) | Patients with KD (n = 46) |
|---|---|---|---|
| Male gender, n (%) | 29 (65.9) | 23 (53.5) | 35 (76.1) |
| Mean (SD), age (y) | 6.0 ± 4.0 | 3.4 ± 2.6 | 1.6 ± 1.5 |
| Age range (y) | 0–16 | 0–16 | 0–9 |
| CAL formation | 22 (47.8%) | ||
| IVIG resistance | 5 (10.9%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, K.-C.; Yang, Y.-L.; Lo, M.-H.; Cai, X.-Y.; Guo, M.M.-H.; Kuo, H.-C.; Huang, Y.-H. Increased Expression of Pyroptosis in Leukocytes of Patients with Kawasaki Disease. Diagnostics 2021, 11, 2035. https://doi.org/10.3390/diagnostics11112035
Kuo K-C, Yang Y-L, Lo M-H, Cai X-Y, Guo MM-H, Kuo H-C, Huang Y-H. Increased Expression of Pyroptosis in Leukocytes of Patients with Kawasaki Disease. Diagnostics. 2021; 11(11):2035. https://doi.org/10.3390/diagnostics11112035
Chicago/Turabian StyleKuo, Kuang-Che, Ya-Ling Yang, Mao-Hung Lo, Xin-Yuan Cai, Mindy Ming-Huey Guo, Ho-Chang Kuo, and Ying-Hsien Huang. 2021. "Increased Expression of Pyroptosis in Leukocytes of Patients with Kawasaki Disease" Diagnostics 11, no. 11: 2035. https://doi.org/10.3390/diagnostics11112035
APA StyleKuo, K.-C., Yang, Y.-L., Lo, M.-H., Cai, X.-Y., Guo, M. M.-H., Kuo, H.-C., & Huang, Y.-H. (2021). Increased Expression of Pyroptosis in Leukocytes of Patients with Kawasaki Disease. Diagnostics, 11(11), 2035. https://doi.org/10.3390/diagnostics11112035







