Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support
Abstract
:1. Introduction
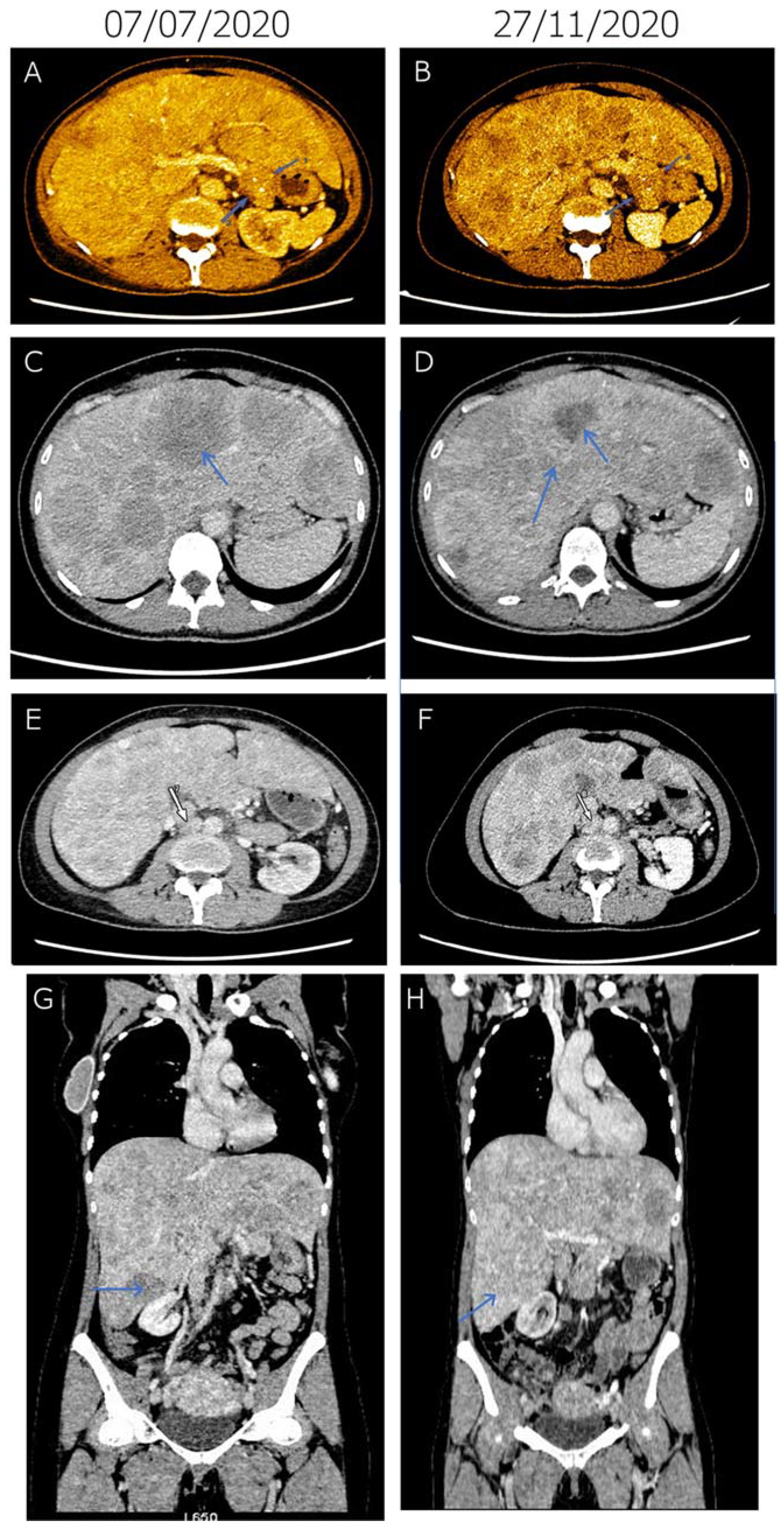
2. Case Report
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Young, K.; Iyer, R.; Morganstein, D.; Chau, I.; Cunningham, D.; Starling, N. Pancreatic neuroendocrine tumors: A review. Futur. Oncol. 2015, 11, 853–864. [Google Scholar] [CrossRef]
- Raymond, E.; Dahan, L.; Raoul, J.-L.; Bang, Y.-J.; Borbath, I.; Lombard-Bohas, C.; Valle, J.; Metrakos, P.; Smith, D.; Vinik, A.; et al. Sunitinib Malate for the Treatment of Pancreatic Neuroendocrine Tumors. N. Engl. J. Med. 2011, 364, 501–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caplin, M.E.; Pavel, M.; Ćwikła, J.B.; Phan, A.T.; Raderer, M.; Sedláčková, E.; Cadiot, G.; Wolin, E.M.; Capdevila, J.; Wall, L.; et al. Lanreotide in Metastatic Enteropancreatic Neuroendocrine Tumors. N. Engl. J. Med. 2014, 371, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Strosberg, J.; El-Haddad, G.; Wolin, E.; Hendifar, A.; Yao, J.; Chasen, B.; Mittra, E.; Kunz, P.L.; Kulke, M.H.; Jacene, H.; et al. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. N. Engl. J. Med. 2017, 376, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.C.; Shah, M.H.; Ito, T.; Bohas, C.L.; Wolin, E.M.; Van Cutsem, E.; Hobday, T.J.; Okusaka, T.; Capdevila, J.; de Vries, E.; et al. Everolimus for Advanced Pancreatic Neuroendocrine Tumors. N. Engl. J. Med. 2011, 364, 514–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, R.V.; Konda, B.; Fountzilas, C.; Mukherjee, S.; Owen, D.; Attwood, K.; Wang, C.; Ma, C.W.; Minderman, H.; Ba, S.S.; et al. Multicenter phase 2 trial of nintedanib in advanced nonpancreatic neuroendocrine tumors. Cancer 2020, 126, 3689–3697. [Google Scholar] [CrossRef]
- Capdevila, J.; Fazio, N.; Lopez, C.L.; Teule, A.; Valle, J.W.; Tafuto, S.; Custodio, A.B.; Reed, N.; Raderer, M.; Grande, E.; et al. Final results of the TALENT trial (GETNE1509): A prospective multicohort phase II study of lenvatinib in patients (pts) with G1/G2 advanced pancreatic (panNETs) and gastrointestinal (giNETs) neuroendocrine tumors (NETs). J. Clin. Oncol. 2019, 37, 4106. [Google Scholar] [CrossRef]
- Xu, J.; Shen, L.; Bai, C.; Wang, W.; Li, J.; Yu, X.; Li, Z.; Li, E.; Yuan, X.; Chi, Y.; et al. Surufatinib in advanced pancreatic neuroendocrine tumours (SANET-p): A randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020, 21, 1489–1499. [Google Scholar] [CrossRef]
- Tirosh, A.; Kebebew, E. Genetic and epigenetic alterations in pancreatic neuroendocrine tumors. J. Gastrointest. Oncol. 2020, 11, 567–577. [Google Scholar] [CrossRef]
- Koschmann, C.; Lowenstein, P.R.; Castro, M.G. ATRX mutations and glioblastoma: Impaired DNA damage repair, alternative lengthening of telomeres, and genetic instability. Mol. Cell. Oncol. 2016, 3, e1167158. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.M.; Rozenblatt-Rosen, O.; Milne, T.; Copeland, T.D.; Levine, S.; Lee, J.C.; Hayes, D.N.; Shanmugam, K.S.; Bhattacharjee, A.; Biondi, C.A.; et al. Menin Associates with a Trithorax Family Histone Methyltransferase Complex and with the Hoxc8 Locus. Mol. Cell 2004, 13, 587–597. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2002, 2, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, L.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Oxnard, G.R.; Paweletz, C.P.; Camidge, D.R.; Heymach, J.V.; Solit, D.B.; Johnson, B.E. Realizing the Potential of Plasma Genotyping in an Age of Genotype-Directed Therapies. J. Natl. Cancer Inst. 2014, 106, dju214. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.; Avila, J.; Rolfo, C.; Ruíz-Patiño, A.; Russo, A.; Ricaurte, L.; Ordóñez-Reyes, C.; Arrieta, O.; Zatarain-Barrón, Z.L.; Recondo, G.; et al. When Tissue is an Issue the Liquid Biopsy is Nonissue: A Review. Oncol. Ther. 2021, 9, 89–110. [Google Scholar] [CrossRef]
- Kamyabi, N.; Bernard, V.; Maitra, A. Liquid biopsies in pancreatic cancer. Expert Rev. Anticancer Ther. 2019, 19, 869–878. [Google Scholar] [CrossRef]
- Lee, J.; Park, S.S.; Lee, Y.K.; Norton, J.A.; Jeffrey, S.S. Liquid biopsy in pancreatic ductal adenocarcinoma: Current status of circulating tumor cells and circulating tumorDNA. Mol. Oncol. 2019, 13, 1623–1650. [Google Scholar] [CrossRef] [Green Version]
- Gall, T.M.; Belete, S.; Khanderia, E.; Frampton, A.E.; Jiao, L.R. Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma. Am. J. Pathol. 2019, 189, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, F.M.; Meyer, T. Liquid Biopsies for Neuroendocrine Tumors: Circulating Tumor Cells, DNA, and MicroRNAs. Endocrinol. Metab. Clin. N. Am. 2018, 47, 471–483. [Google Scholar] [CrossRef]
- Boons, G.; Vandamme, T.; Peeters, M.; Beyens, M.; Driessen, A.; Janssens, K.; Zwaenepoel, K.; Roeyen, G.; Van Camp, G.; De Beeck, K.O. Cell-Free DNA From Metastatic Pancreatic Neuroendocrine Tumor Patients Contains Tumor-Specific Mutations and Copy Number Variations. Front. Oncol. 2018, 8, 467. [Google Scholar] [CrossRef] [Green Version]
- Zakka, K.; Nagy, R.; Drusbosky, L.; Akce, M.; Wu, C.; Alese, O.B.; El-Rayes, B.F.; Kasi, P.M.; Mody, K.; Starr, J.; et al. Blood-based next-generation sequencing analysis of neuroendocrine neoplasms. Oncotarget 2020, 11, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
- Raymond, E.; Thieblemont, C.; Alran, S.; Faivre, S. Impact of the COVID-19 Outbreak on the Management of Patients with Cancer. Target. Oncol. 2020, 15, 249–259. [Google Scholar] [CrossRef]
- Petak, I.; Kamal, M.; Dirner, A.; Bieche, I.; Doczi, R.; Mariani, O.; Filotas, P.; Salomon, A.; Vodicska, B.; Servois, V.; et al. A computational method for prioritizing targeted therapies in precision oncology: Performance analysis in the SHIVA01 trial. NPJ Precis. Oncol. 2021, 5, 1–11. [Google Scholar] [CrossRef]
- Dogruluk, T.; Tsang, Y.H.; Espitia, M.; Chen, F.; Chen, T.; Chong, Z.; Appadurai, V.; Dogruluk, A.; Eterovic, A.K.; Bonnen, P.E.; et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015, 75, 5341–5354. [Google Scholar] [CrossRef] [Green Version]
- Gymnopoulos, M.; Elsliger, M.-A.; Vogt, P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA 2007, 104, 5569–5574. [Google Scholar] [CrossRef] [Green Version]
- Ng, P.K.-S.; Li, J.; Jeong, K.J.; Shao, S.; Chen, H.; Tsang, Y.H.; Sengupta, S.; Wang, Z.; Bhavana, V.H.; Tran, R.; et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell 2018, 33, 450–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dearth, L.R.; Qian, H.; Wang, T.; Baroni, T.E.; Zeng, J.; Chen, S.W.; Yi, S.Y.; Brachmann, R.K. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis 2007, 28, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Jordan, J.J.; Inga, A.; Conway, K.; Edmiston, S.; Carey, L.A.; Wu, L.; Resnick, M.A. Altered-Function p53 Missense Mutations Identified in Breast Cancers Can Have Subtle Effects on Transactivation. Mol. Cancer Res. 2010, 8, 701–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonin-Laurent, N.; Gibaud, A.; Huygue, M.; Lefèvre, S.H.; Le Bras, M.; Chauveinc, L.; Sastre-Garau, X.; Doz, F.; Lumbroso, L.; Chevillard, S.; et al. Specific TP53 mutation pattern in radiation-induced sarcomas. Carcinog 2006, 27, 1266–1272. [Google Scholar] [CrossRef] [Green Version]
- Goldschneider, D.; Horvilleur, E.; Plassa, L.-F.; Guillaud-Bataille, M.; Million, K.; Wittmer-Dupret, E.; Danglot, G.; De Thé, H.; Bénard, J.; May, E.; et al. Expression of C-terminal deleted p53 isoforms in neuroblastoma. Nucleic Acids Res. 2006, 34, 5603–5612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litchfield, K.; Reading, J.L.; Lim, E.L.; Xu, H.; Liu, P.; Al-Bakir, M.; Wong, Y.N.S.; Rowan, A.; Funt, S.A.; Merghoub, T.; et al. Escape from nonsense-mediated decay associates with antitumor immunogenicity. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-D.; Inzunza, H.; Chang, H.; Qi, Z.; Hu, B.; Malone, D.; Cogswell, J. Mutations in the Hedgehog Pathway Genes SMO and PTCH1 in Human Gastric Tumors. PLoS ONE 2013, 8, e54415. [Google Scholar] [CrossRef]
- Martínez-Avilés, L.; Besses, C.; Álvarez-Larrán, A.; Torres, E.; Serrano, S.; Bellosillo, B. TET2, ASXL1, IDH1, IDH2, and c-CBL genes in JAK2- and MPL-negative myeloproliferative neoplasms. Ann. Hematol. 2012, 91, 533–541. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Mullally, A.; Hedvat, C.; Garcia-Manero, G.; Patel, J.; Wadleigh, M.; Malinge, S.; Yao, J.J.; Kilpivaara, O.; Bhat, R.; et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 2009, 114, 144–147. [Google Scholar] [CrossRef]
- Nibourel, O.; Kosmider, O.; Cheok, M.; Boissel, N.; Renneville, A.; Philippe, N.; Dombret, H.; Dreyfus, F.; Quesnel, B.; Geffroy, S.; et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood 2010, 116, 1132–1135. [Google Scholar] [CrossRef]
- Mouliere, F.; Rosenfeld, N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc. Natl. Acad. Sci. USA 2015, 112, 3178–3179. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [Green Version]
- Akirov, A.; Larouche, V.; AlShehri, S.; Asa, S.L.; Ezzat, S. Treatment Options for Pancreatic Neuroendocrine Tumors. Cancers 2019, 11, 828. [Google Scholar] [CrossRef] [Green Version]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; Lazar, V.; Kurzrock, R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J. Clin. Oncol. 2015, 33, 3817–3825. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulou, M.; Stalla, G.K. Somatostatin receptors: From signaling to clinical practice. Front. Neuroendocr. 2013, 34, 228–252. [Google Scholar] [CrossRef]
| Position | Gene Symbol | Protein Alteration | CDS Alteration | dbSNP ID | Driver AEL Score * | Allele Frequency (%) |
|---|---|---|---|---|---|---|
| chr3:179218286 | PIK3CA | p.P539R | c.1616C>G | 121913285 | 70.81 | 65.00 |
| chr17:7675208 | TP53 | p.C135F | c.404G>T | 587781991 | 53.7 | 44.03 |
| chr1:45332803 | MUTYH | p.Y179C | c.536A>G | 34612342 | 12.35 | 89.92 |
| chr16:2064422 | TSC2 | p.E532* | c.1594G>T | 2.68 | 78.11 | |
| chr16:2065544 | TSC2 | p.P542R | c.1625C>G | 764191178 | 2.68 | 91.11 |
| chr3:37047638 | MLH1 | p.K617N | c.1851G>T | 0.61 | 76.68 | |
| chr4:162111346 | FSTL5 | p.E17D | c.51G>C | 140747357 | 0.39 | 46.97 |
| chr6:33320116 | DAXX | p.E454* | c.1360G>T | 0.34 | 80.74 | |
| chr7:99011389 | TRRAP | p.A3717T | c.11149G>A | 199541716 | 0.25 | 45.65 |
| chr20:53576056 | ZNF217 | p.R903Q | c.2708G>A | 61748378 | 0.24 | 47.70 |
| chr2:140475231 | LRP1B | p.A3178T | c.9532G>A | 72899872 | 0.21 | 89.73 |
| chr5:256368 | SDHA | p.L649fs*4 | c.1945_1946delTT | 112307877 | 0.18 | 13.11 |
| chr12:53215425 | RARG | p.R115C | c.343C>T | 0.05 | 2.23 | |
| chr16:56834405 | NUP93 | p.L567P | c.1700T>C | 774760575 | 0.04 | 10.78 |
| chrX:67711447 | AR | p.G644V | c.1931G>T | 0.04 | 78.93 | |
| chr3:138946005 | FOXL2 | p.G240S | c.718G>A | 767088367 | 0.01 | 11.41 |
| chr1:205661956 | SLC45A3 | p.V377M | c.1129G>A | 150525587 | 0 | 88.35 |
| chr10:17071551 | CUBN | p.N834D | c.2500A>G | 759954219 | 0 | 90.93 |
| chr10:86892221 | BMPR1A | p.Q109K | c.325C>A | 0 | 78.77 | |
| chr14:95455532 | SYNE3 | p.R328G | c.982C>G | 145141808 | 0 | 50.75 |
| chr18:33211925 | CCDC178 | p.V737L | c.2209G>C | 117587736 | 0 | 52.54 |
| chr19:40843915 | CYP2A6 | p.V456I | c.1366G>A | 201305272 | 0 | 27.02 |
| chr19:8946760 | MUC16 | p.P10004S | c.30010C>T | 200869910 | 0 | 40.88 |
| chr19:8958984 | MUC16 | p.S5929F | c.17786C>T | 74872724 | 0 | 44.66 |
| chr3:187729913 | BCL6 | p.E164D | c.492G>T | 61752081 | 0 | 91.27 |
| chr3:49897319 | MST1R | p.R715Q | c.2144G>A | 777611015 | 0 | 87.90 |
| chr4:59457 | ZNF595 | p.I11V | c.31A>G | 6834707 | 0 | 16.78 |
| chr5:14291211 | TRIO | p.E346* | c.1036G>T | 0 | 45.38 | |
| chr9:136475373 | SEC16A | p.Y748S | c.2243A>C | 201466249 | 0 | 48.75 |
| chrX:1193297 | CRLF2 | p.K258R | c.773A>G | 1348007359 | 0 | 15.38 |
| chr12:31097896 | DDX11 | p.Q592E | c.1774C>G | 2911826 | −0.01 | 1.99 |
| chr7:129212264 | SMO | p.R726Q | c.2177G>A | 142495470 | −2.5 | 47.40 |
| chr7:108180375 | NRCAM | p.E900G | c.2699A>G | 34721383 | −5 | 51.18 |
| chrX:45063645 | KDM6A | p.T584M | c.1751C>T | 141353229 | −9.33 | 89.78 |
| chr9:8486142 | PTPRD | p.V892A | c.2675T>C | 151005956 | −9.93 | 9.66 |
| chr17:65538235 | AXIN2 | p.S390G | c.1168A>G | 139871607 | −10 | 48.85 |
| chrX:1196817 | CRLF2 | p.V244M | c.730G>A | 151218732 | −24.22 | 100.00 |
| chr4:105234042 | TET2 | p.L34F | c.100C>T | 111948941 | -53.32 | 49.29 |
| Compound | Associated Driver(s) | Compound AEL Score * |
|---|---|---|
| ALPELISIB | PIK3CA p.P539R | 406.80 |
| COPANLISIB | PIK3CA p.P539R | 397.10 |
| EVEROLIMUS | PIK3CA p.P539R TSC2 p.P542R TSC2 p.E532* | 249.20 |
| SIROLIMUS | PIK3CA p.P539R TSC2 p.P542R TSC2 p.E532* | 200.68 |
| BEVACIZUMAB | TP53 p.C135F | 178.03 |
| TEMSIROLIMUS | PIK3CA p.P539R | 171.96 |
| METFORMIN | PIK3CA p.P539R | 171.15 |
| ASPIRIN | PIK3CA p.P539R | 150.57 |
| SUNITINIB | 114.69 | |
| PAZOPANIB | 113.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szkukalek, J.; Dóczi, R.; Dirner, A.; Boldizsár, Á.; Varga, Á.; Déri, J.; Lakatos, D.; Tihanyi, D.; Vodicska, B.; Schwáb, R.; et al. Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support. Diagnostics 2021, 11, 1850. https://doi.org/10.3390/diagnostics11101850
Szkukalek J, Dóczi R, Dirner A, Boldizsár Á, Varga Á, Déri J, Lakatos D, Tihanyi D, Vodicska B, Schwáb R, et al. Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support. Diagnostics. 2021; 11(10):1850. https://doi.org/10.3390/diagnostics11101850
Chicago/Turabian StyleSzkukalek, Judita, Róbert Dóczi, Anna Dirner, Ákos Boldizsár, Ágnes Varga, Júlia Déri, Dóra Lakatos, Dóra Tihanyi, Barbara Vodicska, Richárd Schwáb, and et al. 2021. "Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support" Diagnostics 11, no. 10: 1850. https://doi.org/10.3390/diagnostics11101850
APA StyleSzkukalek, J., Dóczi, R., Dirner, A., Boldizsár, Á., Varga, Á., Déri, J., Lakatos, D., Tihanyi, D., Vodicska, B., Schwáb, R., Pajkos, G., Várkondi, E., Vályi-Nagy, I., Valtinyi, D., Nagy, Z., & Peták, I. (2021). Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support. Diagnostics, 11(10), 1850. https://doi.org/10.3390/diagnostics11101850






