MUTYH as an Emerging Predictive Biomarker in Ovarian Cancer
Abstract
:1. Introduction
2. MUTYH Gene
2.1. MUTYH Gene Function
2.2. MUTYH Germline Mutations
2.3. MUTYH Somatic Mutations
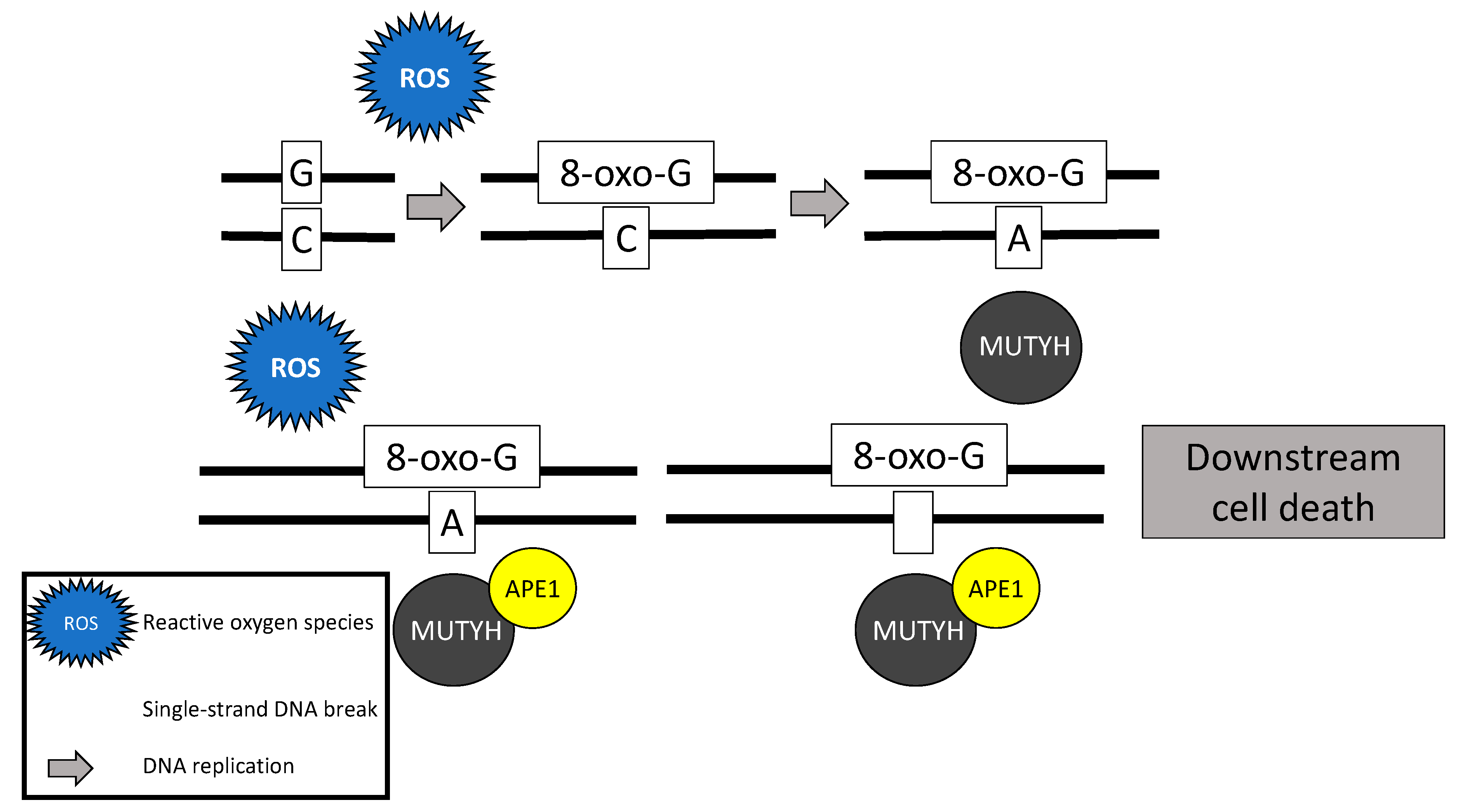
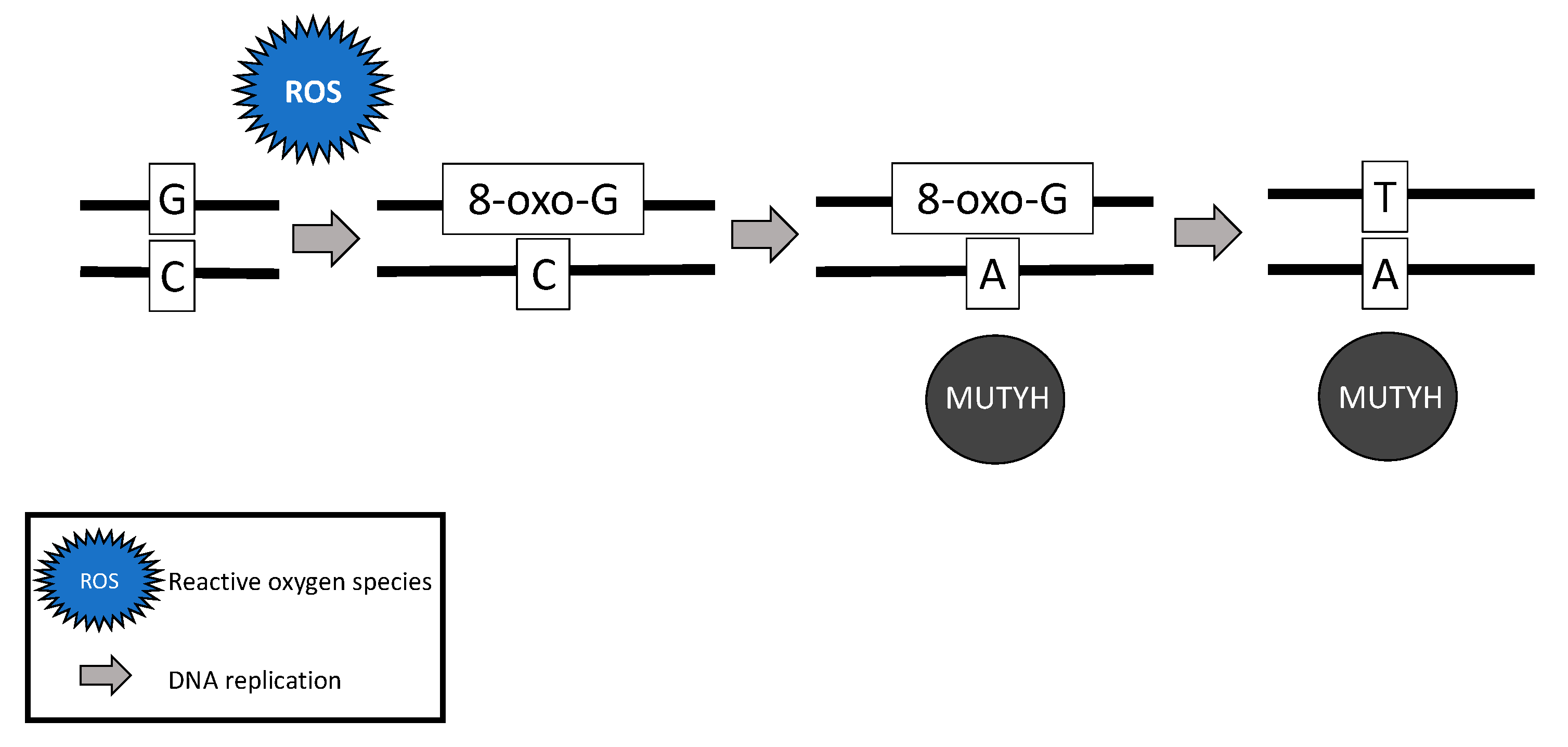
2.4. MUTYH Mutation and Mechanism for Oncogenesis
3. Role of MUTYH in Ovarian Cancer
3.1. Ovarian Cancer Risk
3.2. Early Detection and Prevention of Ovarian Cancer
3.3. Chemotherapeutic Considerations
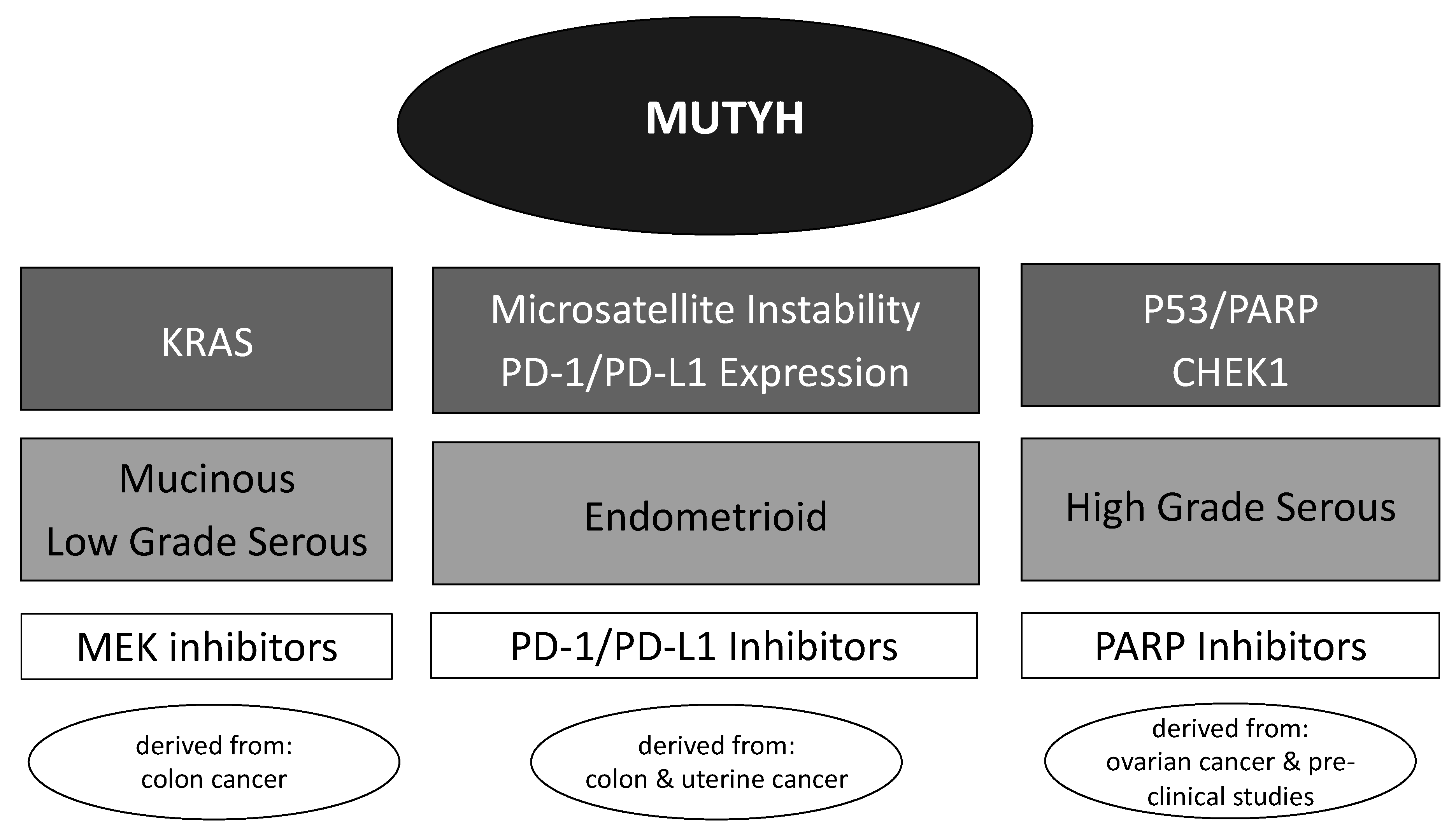
3.4. Potential Targeted Therapeutics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Cancer Institute: Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Ovarian Cancer. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 26 August 2020).
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Royer, R.; Li, S.; McLaughlin, J.R.; Rosen, B.; Risch, H.A.; Fan, I.; Bradley, L.; Shaw, P.A.; Narod, S.A. Frequencies of BRCA1 and BRCA2 mutations among 1342 unselected patients with invasive ovarian cancer. Gynecol. Oncol. 2011, 121, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Schrader, K.A.; Hurlburt, J.; Kalloger, S.E.; Hansford, S.; Young, S.; Huntsman, D.G.; Gilks, C.B.; McAlpine, J.N. Germline BRCA1 and BRCA2 mutations in ovarian cancer: Utility of a histology-based referral strategy. Obstet. Gynecol. 2012, 120, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Graziani, G. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers 2018, 10, 487. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Hart, S.N.; Polley, E.C.; Gnanaolivu, R.; Shimelis, H.; Lee, K.Y.; Lilyquist, J.; Na, J.; Moore, R.; Antwi, S.O.; et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 2018, 319, 2401–2409. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [Green Version]
- Kast, K.; Rhiem, K.; Wappenschmidt, B.; Hahnen, E.; Hauke, J.; Bluemcke, B.; Zarghooni, V.; Herold, N.; Ditsch, N.; Kiechle, M.; et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016, 53, 465–471. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, K.D.; Noltner, K.A.; Buzin, C.H.; Gu, D.; Wen-Fong, C.Y.; Nguyen, V.Q.; Han, J.H.; Lowstuter, K.; Longmate, J.; Sommer, S.S.; et al. Beyond Li Fraumeni Syndrome: Clinical characteristics of families with p53 germline mutations. J. Clin. Oncol. 2009, 27, 1250–1256. [Google Scholar] [CrossRef]
- Kohlmann, W.; Gruber, S.B. Lynch syndrome. In GeneReviews [Internet]; [Updated 2018]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Eds.; University of Washington: Seattle, WA, USA, 2004. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1211/ (accessed on 25 August 2020).
- Møller, P.; Seppälä, T.T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Gareth Evans, D.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.H.; et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut 2018, 67, 1306–1316, Corrected in Gut 2020, 69, e4. [Google Scholar] [CrossRef] [Green Version]
- Nieuwenhuis, M.H.; Vogt, S.; Jones, N.; Nielsen, M.; Hes, F.J.; Sampson, J.R.; Aretz, S.; Vasen, H.F. Evidence for accelerated colorectal adenoma--carcinoma progression in MUTYH-associated polyposis? Gut 2012, 61, 734–738. [Google Scholar] [CrossRef]
- Win, A.K.; Reece, J.C.; Dowty, J.G.; Buchanan, D.D.; Clendenning, M.; Rosty, C.; Southey, M.C.; Young, J.P.; Cleary, S.P.; Kim, H.; et al. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int. J. Cancer 2016, 139, 1557–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, S.; Jones, N.; Christian, D.; Engel, C.; Nielsen, M.; Kaufmann, A.; Steinke, V.; Vasen, H.F.; Propping, P.; Sampson, J.R.; et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 2009, 137, 1976–1985.e10. [Google Scholar] [CrossRef] [PubMed]
- United States National Library of Medicine. MUTYH Gene. Available online: https://medlineplus.gov/genetics/gene/mutyh/ (accessed on 28 July 2020).
- Fry, R.C.; Svensson, J.P.; Valiathan, C.; Wang, E.; Hogan, B.J.; Bhattacharya, S.; Bugni, J.M.; Whittaker, C.A.; Samson, L.D. Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev. 2008, 22, 2621–2626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazouzi, A.; Battistini, F.; Moser, S.C.; Ferreira da Silva, J.; Wiedner, M.; Owusu, M.; Lardeau, C.H.; Ringler, A.; Weil, B.; Neesen, J.; et al. Repair of UV-induced DNA damage independent of nucleotide excision repair is masked by MUTYH. Mol. Cell 2017, 68, 797–807.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molatore, S.; Russo, M.T.; D’Agostino, V.G.; Barone, F.; Matsumoto, Y.; Albertini, A.M.; Minoprio, A.; Degan, P.; Mazzei, F.; Bignami, M.; et al. MUTYH mutations associated with familial adenomatous polyposis: Functional characterization by a mammalian cell-based assay. Hum. Mutat. 2010, 31, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Parker, A.; Wilson, T.M.; Bai, H.; Chang, D.Y.; Lu, A.L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002, 277, 11135–11142. [Google Scholar] [CrossRef] [Green Version]
- Al-Tassan, N.; Chmiel, N.H.; Maynard, J.; Fleming, N.; Livingston, A.L.; Williams, G.T.; Hodges, A.K.; Davies, D.R.; David, S.S.; Sampson, J.R.; et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002, 30, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Raetz, A.G.; David, S.S. When you’re strange: Unusual features of the MUTYH glycosylase and implications in cancer. DNA Repair 2019, 80, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Jansson, K.; Alao, J.P.; Viktorsson, K.; Warringer, J.; Lewensohn, R.; Sunnerhagen, P. A role for Myh1 in DNA repair after treatment with strand-breaking and crosslinking chemotherapeutic agents. Environ. Mol. Mutagen. 2013, 54, 327–337. [Google Scholar] [CrossRef]
- Luncsford, P.J.; Manvilla, B.A.; Patterson, D.N.; Malik, S.S.; Jin, J.; Hwang, B.J.; Gunther, R.; Kalvakolanu, S.; Lipinski, L.J.; Yuan, W.; et al. Coordination of MYH DNA glycosylase and APE1 endonuclease activities via physical interactions. DNA Repair 2013, 12, 1043–1052. [Google Scholar] [CrossRef] [Green Version]
- Oka, S.; Leon, J.; Tsuchimoto, D.; Sakumi, K.; Nakabeppu, Y. MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death. Oncogenesis 2014, 3, e121, Erratum in Oncogenesis 2015, 4, e142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oka, S.; Ohno, M.; Tsuchimoto, D.; Sakumi, K.; Furuichi, M.; Nakabeppu, Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008, 27, 421–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDaid, J.R.; Loughery, J.; Dunne, P.; Boyer, J.C.; Downes, C.S.; Farber, R.A.; Walsh, C.P. MLH1 mediates PARP-dependent cell death in response to the methylating agent N-methyl-N-nitrosourea. Br. J. Cancer 2009, 101, 441–451. [Google Scholar] [CrossRef]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahm, S.H.; Park, J.H.; Ko, S.I.; Lee, Y.R.; Chung, I.S.; Chung, J.H.; Kang, L.W.; Han, Y.S. Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 2011, 44, 352–357. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Piwnica-Worms, H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001, 21, 4129–4139. [Google Scholar] [CrossRef] [Green Version]
- Patil, M.; Pabla, N.; Dong, Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol. Life Sci. 2013, 70, 4009–4021. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.; Infante, E.; Brand, R. MUTYH polyposis. In GeneReviews [Internet]; [Updated 2019]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Eds.; University of Washington: Seattle, WA, USA, 2012. Available online: https://www.ncbi.nlm.nih.gov/books/NBK107219/# (accessed on 30 July 2020).
- Samadder, N.J.; Riegert-Johnson, D.; Boardman, L.; Rhodes, D.; Wick, M.; Okuno, S.; Kunze, K.L.; Golafshar, M.; Uson, P.L.S.; Mountjoy, L.; et al. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol. 2020. [Google Scholar] [CrossRef]
- Win, A.K.; Hopper, J.L.; Jenkins, M.A. Association between monoallelic MUTYH mutation and colorectal cancer risk: A meta-regression analysis. Fam. Cancer 2011, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Win, A.K.; Jenkins, M.A.; Dowty, J.G.; Antoniou, A.C.; Lee, A.; Giles, G.G.; Buchanan, D.D.; Clendenning, M.; Rosty, C.; Ahnen, D.J.; et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2017, 26, 404–412. [Google Scholar] [CrossRef] [Green Version]
- Cheadle, J.P.; Sampson, J.R. MUTYH-associated polyposis--from defect in base excision repair to clinical genetic testing. DNA Repair 2007, 6, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Minion, L.E.; Dolinsky, J.S.; Chase, D.M.; Dunlop, C.L.; Chao, E.C.; Monk, B.J. Hereditary predisposition to ovarian cancer, looking beyond BRCA1/BRCA2. Gynecol. Oncol. 2015, 137, 86–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalogue of Somatic Mutations in Cancer (COSMIC): MUTYH Gene. Available online: https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=MUTYH (accessed on 25 August 2020).
- Bougatef, K.; Marrakchi, R.; Kourda, N.; Ben Lahely, Y.B.; Jileni, S.B.; El Khil, H.K.; Soubrier, F.; Ben Ammar Elgaaied, A. Somatic mutation of MutYH in Tunisian patients with sporadic colorectal cancer. J. Clin. Lab. Anal. 2007, 21, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Parra, G.M.; Gonzalez-Acosta, M.; Thompson, B.A.; Gomez, C.; Fernandez, A.; Damaso, E.; Pons, T.; Morak, M.; Del Valle, J.; Iglesias, S.; et al. Elucidating the molecular basis of MSH2-deficient tumors by combined germline and somatic analysis. Int. J. Cancer 2017, 141, 1365–1380. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, M.L.; Zhao, E.Y.; Reisle, C.; Ch’ng, C.; Wong, H.L.; Shen, Y.; Jones, M.R.; Lim, H.J.; Young, S.; Cremin, C.; et al. Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb. Mol. Case Stud. 2019, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nones, K.; Johnson, J.; Newell, F.; Patch, A.M.; Thorne, H.; Kazakoff, S.H.; de Luca, X.M.; Parsons, M.T.; Ferguson, K.; Reid, L.E.; et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann. Oncol. 2019, 30, 1071–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viel, A.; Bruselles, A.; Meccia, E.; Fornasarig, M.; Quaia, M.; Canzonieri, V.; Policicchio, E.; Urso, E.D.; Agostini, M.; Genuardi, M.; et al. A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine 2017, 20, 39–49. [Google Scholar] [CrossRef]
- Tominaga, Y.; Ushijima, Y.; Tsuchimoto, D.; Mishima, M.; Shirakawa, M.; Hirano, S.; Sakumi, K.; Nakabeppu, Y. MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res. 2004, 32, 3198–3211. [Google Scholar] [CrossRef] [Green Version]
- Rashid, M.; Fischer, A.; Wilson, C.H.; Tiffen, J.; Rust, A.G.; Stevens, P.; Idziaszczyk, S.; Maynard, J.; Williams, G.T.; Mustonen, V.; et al. Adenoma development in familial adenomatous polyposis and MUTYH-associated polyposis: Somatic landscape and driver genes. J. Pathol. 2016, 238, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [Green Version]
- Lefevre, J.H.; Colas, C.; Coulet, F.; Bonilla, C.; Mourra, N.; Flejou, J.F.; Tiret, E.; Bodmer, W.; Soubrier, F.; Parc, Y. MYH biallelic mutation can inactivate the two genetic pathways of colorectal cancer by APC or MLH1 transversions. Fam. Cancer 2010, 9, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Morak, M.; Heidenreich, B.; Keller, G.; Hampel, H.; Laner, A.; de la Chapelle, A.; Holinski-Feder, E. Biallelic MUTYH mutations can mimic Lynch syndrome. Eur. J. Hum. Genet. 2014, 22, 1334–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zachos, G.; Rainey, M.D.; Gillespie, D.A. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003, 22, 713–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurman, R.J.; Shih, I.-M. The Dualistic model of ovarian carcinogenesis: Revisited, revised, and expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudson, A.G., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, K.; Berek, J.S.; Chen, L.-M.; Cristea, M.; DeRosa, M.; et al. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology: Ovarian Cancer Including Fallopian Tube Cancer and Primary Peritoneal Cancer. Version 1. 2020. Available online: https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf (accessed on 26 August 2020).
- Konstantinopoulos, P.A.; Lacchetti, C.; Annunziata, C.M. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline summary. JCO Oncol. Pract. 2020, 16, e835–e838. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, J.T.; Louie, E.W.; Pike, M.C.; Roy, S.; Ross, R.K.; Henderson, B.E. “Incessant ovulation” and ovarian cancer. Lancet 1979, 2, 170–173. [Google Scholar] [CrossRef]
- Cramer, D.W.; Welch, W.R. Determinants of ovarian cancer risk. II. Inferences regarding pathogenesis. J. Natl. Cancer Inst. 1983, 71, 717–721. [Google Scholar]
- Medeiros, F.; Muto, M.G.; Lee, Y.; Elvin, J.A.; Callahan, M.J.; Feltmate, C.; Garber, J.E.; Cramer, D.W.; Crum, C.P. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am. J. Surg. Pathol. 2006, 30, 230–236. [Google Scholar] [CrossRef]
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian cancers: Genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization: International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Internal Report 14/002: Report of the Advisory Group to Recommend Priorities for IARC Monographs during 2015–2019. Available online: https://monographs.iarc.fr/wp-content/uploads/2018/08/14-002.pdf (accessed on 28 October 2020).
- Pingarilho, M.; Oliveira, N.G.; Martins, C.; Fernandes, A.S.; de Lima, J.P.; Rueff, J.; Gaspar, J.F. Genetic polymorphisms in detoxification and DNA repair genes and susceptibility to glycidamide-induced DNA damage. J. Toxicol. Environ. Health A 2012, 75, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Nagata, C.; Konishi, K.; Tamura, T.; Wada, K.; Tsuji, M.; Hayashi, M.; Takeda, N.; Yasuda, K. Associations of acrylamide intake with circulating levels of sex hormones and prolactin in premenopausal Japanese women. Cancer Epidemiol. Biomarkers Prev. 2015, 24, 249–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, M.I.; El-Demerdash, F.M. Acrylamide-induced oxidative stress and biochemical perturbations in rats. Toxicology 2006, 219, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Adani, G.; Filippini, T.; Wise, L.A.; Halldorsson, T.I.; Blaha, L.; Vinceti, M. Dietary intake of acrylamide and risk of breast, endometrial, and ovarian cancers: A systematic review and dose-response meta-analysis. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1095–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Miron, A.; Drapkin, R.; Nucci, M.R.; Medeiros, F.; Saleemuddin, A.; Garber, J.; Birch, C.; Mou, H.; Gordon, R.W.; et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J. Pathol. 2007, 211, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Soong, T.R.; Howitt, B.E.; Miron, A.; Horowitz, N.S.; Campbell, F.; Feltmate, C.M.; Muto, M.G.; Berkowitz, R.S.; Nucci, M.R.; Xian, W.; et al. Evidence for lineage continuity between early serous proliferations (ESPs) in the Fallopian tube and disseminated high-grade serous carcinomas. J. Pathol. 2018, 246, 344–351. [Google Scholar] [CrossRef]
- Clarke-Pearson, D.L. Clinical practice. Screening for ovarian cancer. N. Engl. J. Med. 2009, 361, 170–177. [Google Scholar] [CrossRef]
- McLaughlin, J.R.; Risch, H.A.; Lubinski, J.; Moller, P.; Ghadirian, P.; Lynch, H.; Karlan, B.; Fishman, D.; Rosen, B.; Neuhausen, S.L.; et al. Reproductive risk factors for ovarian cancer in carriers of BRCA1 or BRCA2 mutations: A case-control study. Lancet Oncol. 2007, 8, 26–34. [Google Scholar] [CrossRef]
- Michels, K.A.; Pfeiffer, R.M.; Brinton, L.A.; Trabert, B. Modification of the associations between duration of oral contraceptive use and ovarian, endometrial, breast, and colorectal cancers. JAMA Oncol. 2018, 4, 516–521. [Google Scholar] [CrossRef]
- Daly, M.B.; Pal, T.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Goggins, M.; Hutton, M.L.; et al. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. Version 2. 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (accessed on 30 September 2020).
- Skates, S.J.; Greene, M.H.; Buys, S.S.; Mai, P.L.; Brown, P.; Piedmonte, M.; Rodriguez, G.; Schorge, J.O.; Sherman, M.; Daly, M.B.; et al. Early detection of ovarian cancer using the Risk of Ovarian Cancer Algorithm with frequent CA125 testing in women at increased familial risk—Combined results from two screening trials. Clin. Cancer Res. 2017, 23, 3628–3637. [Google Scholar] [CrossRef] [Green Version]
- Gaillard, S.L.; Bookman, M.A. Principles of chemotherapy in gynecologic cancer. In Principles and Practice of Gynecologic Oncology, 7th ed.; Chi, D.S., Berchuck, A., Dizon, D.S., Yashar, C., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2017; pp. 275–302. [Google Scholar]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Jia, Y.; Wang, S.; Liu, N.; Gao, D.; Zhang, L.; Lin, Z.; Wang, S.; Kong, F.; Peng, C.; et al. Downregulation of MUTYH contributes to cisplatin resistance of esophageal squamous cell carcinoma cells by promoting Twist mediated EMT. Oncol. Rep. 2019, 42, 2716–2727. [Google Scholar] [CrossRef] [PubMed]
- Aebi, S.; Kurdi-Haidar, B.; Gordon, R.; Cenni, B.; Zheng, H.; Fink, D.; Christen, R.D.; Boland, C.R.; Koi, M.; Fishel, R.; et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996, 56, 3087–3090. [Google Scholar] [PubMed]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Chang, D.Y.; Lu, A.L. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005, 280, 408–417. [Google Scholar] [CrossRef] [Green Version]
- Friedrich-Heineken, E.; Toueille, M.; Tännler, B.; Bürki, C.; Ferrari, E.; Hottiger, M.O.; Hübscher, U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005, 353, 980–989. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Chua, J.K.; Seah, K.S.; Koo, S.H.; Yee, J.Y.; Yang, E.G.; Lim, K.K.; Pang, S.Y.; Yuen, A.; Zhang, L.; et al. Predicting chemotherapeutic drug combinations through gene network profiling. Sci. Rep. 2016, 6, 18658. [Google Scholar] [CrossRef] [Green Version]
- Cass, I.; Baldwin, R.L.; Varkey, T.; Moslehi, R.; Narod, S.A.; Karlan, B.Y. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 2003, 97, 2187–2195. [Google Scholar] [CrossRef]
- Sakai, W.; Swisher, E.M.; Karlan, B.Y.; Agarwal, M.K.; Higgins, J.; Friedman, C.; Villegas, E.; Jacquemont, C.; Farrugia, D.J.; Couch, F.J.; et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008, 451, 1116–1120. [Google Scholar] [CrossRef] [Green Version]
- Hussain, T.; Mulherkar, R. Lymphoblastoid cell lines: A continuous in vitro source of cells to study carcinogen sensitivity and DNA repair. Int. J. Mol. Cell Med. 2012, 1, 75–87. [Google Scholar]
- Nielsen, M.; de Miranda, N.F.; van Puijenbroek, M.; Jordanova, E.S.; Middeldorp, A.; van Wezel, T.; van Eijk, R.; Tops, C.M.; Vasen, H.F.; Hes, F.J.; et al. Colorectal carcinomas in MUTYH-associated polyposis display histopathological similarities to microsatellite unstable carcinomas. BMC Cancer 2009, 9, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Dong, D.; He, W.; Song, L.; Wang, Q.; Yue, J.; Xie, L. Mismatch repair deficiency is associated with MSI phenotype, increased tumor-infiltrating lymphocytes and PD-L1 expression in immune cells in ovarian cancer. Gynecol. Oncol. 2018, 149, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Vaddepally, R.K.; Kharel, P.; Pandey, R.; Garje, R.; Chandra, A.B. Review of Indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers 2020, 12, 738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boland, P.M.; Yurgelun, M.B.; Boland, C.R. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 2018, 68, 217–231. [Google Scholar] [CrossRef]
- Volkov, N.M.; Yanus, G.A.; Ivantsov, A.O.; Moiseenko, F.V.; Matorina, O.G.; Bizin, I.V.; Moiseyenko, V.M.; Imyanitov, E.N. Efficacy of immune checkpoint blockade in MUTYH-associated hereditary colorectal cancer. Investig. New Drugs 2020, 38, 894–898. [Google Scholar] [CrossRef]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Miao, Z.H.; Zhu, H.; Cai, Y.J.; Lu, W.; Ding, J. Chk1 and Chk2 are differentially involved in homologous recombination repair and cell cycle arrest in response to DNA double-strand breaks induced by camptothecins. Mol. Cancer Ther. 2008, 7, 1440–1449. [Google Scholar] [CrossRef] [Green Version]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [Green Version]
- Amado, R.G.; Wolf, M.; Peeters, M.; Van Cutsem, E.; Siena, S.; Freeman, D.J.; Juan, T.; Sikorski, R.; Suggs, S.; Radinsky, R.; et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 1626–1634. [Google Scholar] [CrossRef]
- Benson III, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Garrido-Laguna, I.; et al. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology: Colon Cancer. Version 4. 2020. Available online: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf (accessed on 30 October 2020).
- Jänne, P.A.; Shaw, A.T.; Pereira, J.R.; Jeannin, G.; Vansteenkiste, J.; Barrios, C.; Franke, F.A.; Grinsted, L.; Zazulina, V.; Smith, P.; et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: A randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013, 14, 38–47. [Google Scholar] [CrossRef]
- Nakayama, N.; Nakayama, K.; Yeasmin, S.; Ishibashi, M.; Katagiri, A.; Iida, K.; Fukumoto, M.; Miyazaki, K. KRAS or BRAF mutation status is a useful predictor of sensitivity to MEK inhibition in ovarian cancer. Br. J. Cancer 2008, 99, 2020–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, K.; McAlpine, J.N.; Lheureux, S.; Matsumura, N.; Oza, A.M. Paradigm Shift in the Management Strategy for Epithelial Ovarian Cancer. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e247–e257. [Google Scholar] [CrossRef] [PubMed]
| Cancer Type | Risk in Monoallelic Carriers | Risk in Biallelic Carriers |
|---|---|---|
| Colon Cancer | Possible increased risk | 63% 1 |
| Bladder Cancer | Insufficient evidence | 25% 2 (males) 8% 2 (females) |
| Ovarian Cancer | No increased risk | 14% 2 (females) |
| Duodenal Cancer | Insufficient evidence | 4% 3 |
| Breast Cancer | 11% 1 (females) | 25% 3 (females) |
| Gastric Cancer | 5% 1 (males) 2.3% 1 (females) | Insufficient evidence |
| Hepatobiliary Cancer | 3% 1 (males) 1.4% 1 (females) | Insufficient evidence |
| Endometrial Cancer | 3% 1 (females) | Possible increased risk |
| Skin Cancer | No increased risk | Possible increased risk |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hutchcraft, M.L.; Gallion, H.H.; Kolesar, J.M. MUTYH as an Emerging Predictive Biomarker in Ovarian Cancer. Diagnostics 2021, 11, 84. https://doi.org/10.3390/diagnostics11010084
Hutchcraft ML, Gallion HH, Kolesar JM. MUTYH as an Emerging Predictive Biomarker in Ovarian Cancer. Diagnostics. 2021; 11(1):84. https://doi.org/10.3390/diagnostics11010084
Chicago/Turabian StyleHutchcraft, Megan L., Holly H. Gallion, and Jill M. Kolesar. 2021. "MUTYH as an Emerging Predictive Biomarker in Ovarian Cancer" Diagnostics 11, no. 1: 84. https://doi.org/10.3390/diagnostics11010084
APA StyleHutchcraft, M. L., Gallion, H. H., & Kolesar, J. M. (2021). MUTYH as an Emerging Predictive Biomarker in Ovarian Cancer. Diagnostics, 11(1), 84. https://doi.org/10.3390/diagnostics11010084







