Abstract
The discovery of non-coding RNAs (ncRNAs) has opened a new paradigm to use ncRNAs as biomarkers to detect disease progression. Long non-coding RNAs (lncRNA) have garnered the most attention due to their specific cell-origin and their existence in biological fluids. Type 2 diabetes patients will develop cardiovascular disease (CVD) complications, and CVD remains the top risk factor for mortality. Understanding the lncRNA roles in T2D and CVD conditions will allow the future use of lncRNAs to detect CVD complications before the symptoms appear. This review aimed to discuss the roles of lncRNAs in T2D and CVD conditions and their diagnostic potential as molecular biomarkers for CVD complications in T2D.
1. Introduction
Type 2 diabetes (T2D) is a disease of high blood glucose (hyperglycemia) due to insulin action failure. About 463 million individuals have diabetes worldwide, with 90% of them T2D patients [1]. The number is going to increase to 700 million affected people worldwide in 2045 [1]. The increasing prevalence of diabetes worldwide is a burden, as most T2D patients will develop macrovascular and microvascular complications, which lead to various severe health problems and mortality [2,3,4]. Despite advances and improvements in the treatment options for T2D, the majority of deaths in T2D patients are due to cardiovascular diseases (CVD) [5,6,7,8]. People with diabetes have a CVD mortality risk 3- to 4-fold higher than those without diabetes [6,9]. Both T2D and CVD share some pathophysiological pathways and risk factors, particularly in the development of vascular damage, causing atherosclerosis [2,4,10]. These pathophysiologic changes will be elaborated in the next section. Early identification of which diabetic patients will develop CVD is important to reduce the severe disease outcomes and mortality.
The mapping of the whole human genome and transcriptome showed that the protein-coding genes account for only 2% of the whole human genome, where the rest consist of undefined or non-coding regions [11,12]. Recent evidence showed that 98% of the genome are the functional non-coding RNAs [12,13,14,15]. Non-coding RNAs (ncRNAs) are grouped into their sizes: (1) the small ncRNAs (size <200 bp) such as microRNA (miRNA) and some of the circular RNA (circRNA), and (2) the large ncRNAs such as long non-coding RNA (lncRNA) [13,14,15]. Among these ncRNAs, miRNA is most studied and comprehensively reviewed in T2D and CVD development [16,17,18], as well as being potential therapeutic targets [19,20,21]. In comparison, research on lncRNA and circRNA in T2D or CVD is still limited [22,23,24,25]. Previous reviews have discussed the interactions of these ncRNAs (miRNA-lncRNA-circRNA) in contributing to CVD development [26,27,28] and diabetes-related diseases [29,30,31]. Since lncRNAs are stably present in the biological fluids [32], they have similar potential to miRNA as new biomarkers or therapeutic targets. This review aimed to summarize the recent findings of circulating lncRNAs as the potential biomarkers for CVD complications in T2D. How these lncRNAs are involved in CVD development in T2D will also be explored.
2. Long Non-Coding RNAs (lncRNAs) in Type 2 Diabetes (T2D)
Long non-coding RNAs (lncRNAs) are the RNA molecules with a size of more than 200 nucleotides and without the protein translation capacity [33,34]. The biogenesis and functions of lncRNAs have been discussed extensively [33,34]. LncRNAs are heterogeneous due to their genomic origins, in which they arise from the unconserved regions of the genome [33,34]. They are similar to messenger RNAs in their primary structure, with some of the lncRNAs having a 3′polyadenylation tail, although this 3′tail is not necessarily for their functions [35]. For the lncRNAs’ secondary and tertiary structures, the structural motifs are relatively conserved and vital for their functions [36,37]. About 27,919 human lncRNAs have been reported, and 70% of them are functional [38]. LncRNAs regulate the transcription process by interacting with the proteins, RNA, and DNA molecules in the cis- and trans-regulation activities [33,34]. In the cis-regulation, lncRNAs act on the neighbouring genes on the same allele of their origin. In the trans-regulation, lncRNAs exert their regulatory activities on distant locations from their origin [33,34]. Among the reported regulatory functions, lncRNAs interact with the chromatin-modifying complexes to facilitate the chromatin remodeling and initiate the transcription of the downstream genes [33,34]. LncRNAs also act as scaffolds to stabilize complexes, allow protein–protein or protein–RNA formations, and direct these complexes to specific locations on the DNA for the intended regulation [33,34]. Another known function of lncRNA is to act as a microRNA (miRNA) sponge, in which lncRNA binds to the miRNA to prevent the target gene silencing [33,34]. Previous GWAS studies identified genetic variants in the lncRNA antisense non-coding RNA gene at the INK4 locus (ANRIL) that was associated with a higher risk of having T2D [39] and coronary artery disease (CAD) [40], therefore, suggesting the involvement of lncRNAs in diabetic CVD complication. First, we summarized and discussed the reported lncRNAs involved in the T2D environment (Table 1).

Table 1.
Summary of the reported long non-coding RNAs (lncRNAs) in Type 2 Diabetes (T2D). List of previously published lncRNAs in T2D environment and their respective information.
2.1. LncRNA in Insulin Secretion
Dysregulation of the lncRNAs involved in the pancreatic β-cell development and function could contribute to the failure of or reduced insulin secretion (Table 1 and Figure 1) [71]. One of the main events in T2D disease development is the failure of the pancreatic β-cell compensatory insulin secretion in response to insulin resistance [72]. The increase of the insulin secretion depends on the ability of each β-cells to produce more insulin (β-cell function per cell) or high numbers of existing β-cells in the islet (β-cell mass) [72]. Therefore, dysregulated lncRNAs in the islet and β-cells development will cause insulin secretion failure. One such lncRNA is the β-cell long intergenic non-coding RNA 1 (βlinc1), a conserved lncRNA located upstream of the NK2 homeobox 2 (NKX2-2) gene (an essential islet transcription factor) [73]. A knockout mouse model study revealed that lncRNA βlinc1 is necessary for proper coordinated islet-specific transcription factors [44]. Deleting βlinc1 in the mouse model causes defective islets and disrupts the glucose homeostasis in the adult knockout mice [44]. Transcriptomic profiling of human islets revealed that some lncRNAs are exclusively expressed in the islets [41]. One particular lncRNA is the human islet long non-coding RNA 25 (HI-LNC25) that is located near to MAF bZIP transcription factor B (MAFB) gene (an essential gene for islet maturation), and this lncRNA regulates the expression of GLIS family zinc finger 3 (GLIS3, islet transcription factor) [41]. In the same study [41], the expressions of lncRNA KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) and HI-LNC45 lncRNAs were upregulated and downregulated, respectively, in T2D patients. Although their functions are unknown, lncRNA KCNQ1OT1 is one of the T2D susceptible loci [74]. Other reported islet-specific lncRNAs are the HI-LNC12, HI-LNC78, and HI-LNC71, in which the knockdown of these three lncRNAs caused impaired insulin secretion [49]. Importantly, lncRNA HI-LNC71 is located upstream of the pancreatic and duodenal homeobox 1 (PDX1) gene in an antisense orientation, and a knockdown of lncRNA HI-LNC71 in the mice reduced the expression of PDX1. Since PDX1 is the main regulator of pancreatic development and β-cell function, the loss of lncRNA HI-LNC71 may indeed cause disruption in pancreatic function and subsequently induce T2D [49]. A more recent study also reported another lncRNA, LINC00261, that is also important for the islet differentiation, in which it interacts with the islet transcription factor, forkhead box A2 (FOXA2) [75]. These findings suggest that most of these lncRNAs are from the regions near the essential islet differentiation and development factors, indicating the regulatory role of these lncRNAs during the gene transcription process.
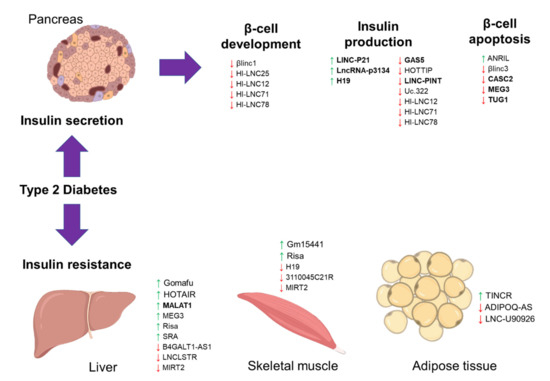
Figure 1.
The schematic diagram illustrates the long noncoding RNA (lncRNA) involvement in Type 2 diabetes (T2D). The diagram shows the involvement of previously published long noncoding RNAs (lncRNAs) in T2D, focusing on insulin secretion and resistance. A green upward arrow indicates the upregulation of the lncRNA expression, a red downward arrow indicates the reduced expression, and the bold lncRNAs exist in the circulating biological samples for diagnostic potential.
In terms of β-cell insulin production and secretion, lncRNA growth arrest-specific 5 (GAS5) is the most reported lncRNA in the association with T2D. In a study of the T2D mouse model, lncRNA GAS5 expression was lower in the islets of T2D mice, and silencing of this lncRNA expression caused impaired insulin synthesis and secretion in the MIN6 cells [46]. A similar observation was seen in a study of T2D patients, in which the circulating expression of GAS5 was reduced in T2D patients [47]. Silencing of GAS5 expression in the rat islet cell line reduced the expression of multiple microRNAs (miR-29a-3p, miR-96-3p, and miR-208a-3p) and subsequently reduced these microRNAs’ targets expressions, including the insulin receptor, insulin receptor substrate, and phosphoinositide-3-kinase regulatory subunit-1 [47]. Transcriptomic profiling of the human islets also found LOC283177 is co-expressed with the insulin synthesis and secretion regulators, including MAP-kinase activating death domain (MADD), synaptotagmin 11 (SYT11), and paired box 6 (PAX6) [76]. Besides that, lncRNA uc.322 is also exclusively expressed in pancreatic β-cells [55]. Overexpression of lncRNA uc.322 in the MIN6 cells caused greater glucose-stimulated insulin secretion via the upregulations of the PDX1 and forkhead box O1 (FOXO1) expressions [55]. Another lncRNA is the long intergenic non-protein coding RNA, p53 induced transcript (LINC-PINT), that may regulate insulin synthesis and secretion, and this lncRNA expression was reduced in the T2D mouse model [51] and plasma of T2D patients [77]. In contrast, expression of LINC-P21 was higher in the serum of T2D patients, and silencing of this lncRNA resulted in better glucose-stimulated insulin secretion of the INS-1 cells via miR-766-3p/NR3C2 axis [42]. Recent profiling of T2D patients showed that serum exosomal lncRNA-p3134 was enriched in T2D patients and correlated with fasting blood glucose and HOMA-β levels [43]. This enrichment of lncRNA-p3134 expression was only seen in serum exosomes, not in the serum samples alone, indicating a specific enrichment in the extracellular vesicles. Also, lncRNA-p3134 regulates insulin secretion by promoting the expressions of critical modulators (PDX1, MAFA, GLUT2, and TCF7L2), possibly via the PI3K/Akt/mTOR signaling in the pancreatic β cells [43].
The progressive loss of β-cells is common in T2D disease development [78]. LncRNA is also implicated in β-cell death. A previous study of human and mice islets showed that lncRNA βlinc3 expression was low in T2D condition, and this lncRNA promotes β-cell apoptosis [45]. Another reported lncRNA is taurine up-regulated 1 (TUG1) that also plays a role in β-cells, and a reduced expression of TUG1 causes apoptosis and impaired insulin secretion [54]. Maternally expressed 3 (MEG3), a maternally imprinted lncRNA, was also reduced in the islets of T2D patients [52]. This reduced expression was due to hypermethylation on the cluster region of MEG3 location, caused the suppression of the same cluster’s microRNAs [52]. Functional analysis of these dysregulated microRNAs showed that they are involved in β-cell apoptosis and T2D pathogenesis [52], which may imply that MEG3 could also regulate β-cell function. Further investigation was done in the T2D mouse model, in which silencing of MEG3 expression led to impaired insulin production and β-cell apoptosis [53]. Circulating lncRNA H19 imprinted maternally expressed transcript (H19) was reduced in T2D patients, and this low expression of lncRNA H19 was associated with the increase of miR-29a and miR29b expressions [48]. High expression of the miR-29 family causes β-cell death and dysfunction [79], thus may imply that lncRNA H19 presence is important for normal β-cell function. This H19 role is supported by another study of mice islets, in which high expression H19 promoted the expansion of β-cell mass in response to increasing glucose demand [80]. Another is the lncRNA HOXA distal transcript antisense RNA (HOTTIP), which was lower in the T2D mouse model, and silencing of this lncRNA caused reduced insulin secretion and its regulators expressions (PDX1 and MAFA) [50]. In-depth investigation showed that lncRNA HOTTIP also regulates cyclin proteins and β-cell cycle [50]. These interactions between the lncRNA and microRNAs may explain how the loss of these lncRNAs could contribute to dysregulation of the insulin secretion vital regulators.
2.2. LncRNA in Insulin Resistance
Insulin resistance is due to the impaired response of insulin-responsive cells, and lncRNA may regulate insulin signaling (Table 1 and Figure 1). One such lncRNA is MEG3 that was higher in the liver of the high-fat-fed and obese mouse models [59]. High MEG3 expression causes hepatic insulin resistance by increasing the expression of FOXO1, which then promotes hepatic gluconeogenesis. Notably, the increase of MEG3 expression due to high-fat diets caused increased histone acetylation, removing the epigenetic suppression on the MEG3 cluster [59]. Similarly, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was also increased in the obese mouse model’s liver and promoted insulin resistance [58]. MALAT1 stabilizes the sterol regulatory element-binding transcription factor 1c (SREBP-1c) in the nucleus and causes excess lipid accumulation in the hepatocytes [58]. Another reported lncRNA contributing to hepatic lipid accumulation is the lncRNA steroid receptor RNA activator (SRA). LncRNA SRA suppresses adipose triglyceride lipase (ATGL) expression and inhibits fatty acid oxidation, causing hepatic lipid accumulation [61]. A protective liver lncRNA, liver-specific triglyceride regulator (LNCLSTR), was shown to positively regulate plasma triglyceride clearance via interacting with the TDP-43/FXR/APOC2 pathway [63], thus may be beneficial for treating insulin resistance. A recent review has discussed these lncRNAs in-depth and discussed their potential as early diagnostic markers for insulin resistance [81]. However, many of these lncRNAs are also present for other metabolic diseases such as obesity and fatty liver; thus, their roles are not specific to the T2D environment.
2.3. Circulating lncRNAs as Biomarkers for T2D
Some of these lncRNAs are present in the circulating biological fluids, and thus some studies determined the potential of these lncRNAs as biomarkers for T2D (Table 2). One such study is the cohort of US army veterans, in which the serum lncRNA GAS5 level was lower in the T2D patients, and those with serum GAS5 level lower than 10 ng/µL have 12 times greater risk to have T2D [82]. Moreover, the diagnostic analysis using the Receiver operating characteristic (ROC) analysis showed that serum GAS5 level could differentiate the T2D patients from the healthy controls with an area under the curve (AUC) of ROC of 0.81, with 67.3% specificity and 85.1% sensitivity [82]. A recent systematic review reported that out of the seven studies, circulating expression of lncRNAs could differentiate the individuals with T2D from the healthy controls with a minimal sensitivity (0.71) and specificity (0.66) [83]. A total of 11 blood lncRNAs were identified from the previous studies, in which three lncRNAs were upregulated (ENST00000550337.1, TCONS_00007244, and TCONS_00000886 [84]), and eight lncRNAs were downregulated (VIM-AS1, CTBP1-AS2 [85], LY86-AS1, HCG27_201 [86], LINC00523, LINC00994 [87], CASC2 [88], and GAS5 [82,89]) in T2D individuals. In terms of genetic susceptibility of T2D, lncRNA ANRIL is consistently reported to increase the risk of having T2D and CAD [90]. In this locus, the genetic alterations are different for T2D and CAD, in which most of the T2D genetic risk variants are located distal to the last exon, except one variant (rs564398) that is associated with both CAD and T2D conditions [91]. Although the mechanism of how ANRIL regulates glucose metabolism and insulin secretion is unknown, the circulating expression of ANRIL was higher in T2D patients, patients with myocardial infarction (MI) only, and much higher in T2D patients with MI, thus indicating the role of ANRIL in CVD progression of T2D patients [92]. Furthermore, identification of the role of these T2D-lncRNAs may provide a better understanding of the molecular network in the development of T2D. As recently reported, the treatment of glucagon-like peptide-1 receptor agonists (GLP1RAs) in the T2D cell model showed that lncRNA NONRATT027738 is in the core of the molecular networks that resulted in the β-cell improvements following the GLP1ARs treatment [93].

Table 2.
Summary of the reported circulating long non-coding RNAs (lncRNAs) as biomarkers for Type 2 diabetes (T2D). List of previously published circulating lncRNAs as biomarkers for T2D and their respective information.
3. Cardiovascular Disease (CVD) Complication in T2D
CVD encompasses the anomalies in the heart and vascular circulatory system, including myocardial ischemia/injury and infarction (MI), atherosclerosis, hypertension, myocarditis, valvular disease, coronary artery disease, and arrhythmias [2,4]. With other risk factors such as obesity, aging, and diabetes, these anomalies eventually lead to heart failure (HF) [2,4]. There is a significant gap in understanding the mechanisms and the signaling pathways involved in these CVD events in T2D. What is known is that the pathophysiology of the CVD complication in T2D depends on the balance between the damage or dysfunctional changes that occur due to the T2D environment and the response of the endogenous protective factors of the heart [2]. These changes due to T2D in the vascular system and blood cause hemodynamic stress on the heart [2]. As a response, the heart or endothelial cells make a series of genetic, molecular, and cellular changes known as the cardiac remodeling process, including cardiac hypertrophy, fibrosis, inflammation, and vascular changes [94]. Thus, depending on T2D individuals, their physiology’s functional changes at the microvascular level can differ from at the macrovascular level. Therefore, they may vary the implications with respect to future CVD risk and outcomes.
In the T2D environment, one of the early critical events for CVD complications is the increase in arterial stiffness and endothelial cell damage that contributes to atherosclerosis [95,96]. The high levels of circulating glucose and lipids are significant determinants for both arterial stiffness and carotid intimal media thickness (IMT) [97,98], and this association is only evident in T2D patients [99]. Thus, further confirming that there are specific metabolic changes in the T2D environment in those with CVD complications. The elevated circulating glucose and triglycerides, particularly the low-density lipoprotein (LDL), are taken in by the arterial intima (a layer of the endothelial cells in the artery) and consequently, recruits the macrophages to the site (an early formation of the atherosclerotic plaque), to take up the lipoproteins and causes their differentiation into the foam cells [100].
Eventually, this event leads to high production of inflammatory cytokines and chemokines together with excessive release of the reactive oxygen species (ROS) and AGEs, thus activating the nuclear factor-κB (NFκB), activator protein-1 (AP-1), endothelin-1 (ET-1, a vasoconstrictive signal), pro-thrombotic tissue factors, and plasminogen activator inhibitor-1 (PAI-1) [100,101,102]. The increased levels of these molecules contribute to inflammation, vasoconstriction, and thrombosis [100,101,102]. Additionally, the presence of insulin resistance also contributes to greater ROS production [103]. ROS activates higher expressions of adhesion molecules in endothelial cells, which further recruits more leukocytes into the plaque and increases the formation of the extracellular matrix (ECM) in this area [100,103]. Similar to insulin resistance, disruption of protein kinase C action also contributes to impaired angiogenesis, vasodilation, and greater leukocyte recruitment to the plaque by inhibiting PI3K signal [100]. This atherosclerotic plaques block the blood flow and lead to life-threatening events such as myocardial infarction (MI) and stroke [100].
As a response to these changes within the T2D environment, cardiac remodeling, which is defined by genetic, molecular, cellular changes that result in changes in the shape, size, and function of the cardiac cells, will occur [94]. The failure of this remodeling can result in cardiac dysfunction or, in the worst cases, heart failure (HF) [104]. Although the exact mechanisms or effects of T2D on cardiac remodeling are unknown, evidence of either dilated or restrictive left ventricular (LV) phenotypes is seen in T2D patients [105]. One of the reported failed cardiac remodeling mechanisms is where the heart activates the fetal expression of cardiac genes that causes the loss of ventricular function [94]. This event is asymptomatic at the start but eventually will lead to HF [104]. Another mechanism is the ion channel remodeling, such as the inactivation of the sodium channels, changes in calcium and potassium channels resulting in ventricular arrhythmias [106]. These ion changes impair the cellular contractility and prolong the cardiac action potential (AP) due to the reduced membrane repolarizing current [106].
Another cause for HF is the reduced expression of Connexin 43 (CX43) [107]. CX43 is the most highly expressed gap junction protein in the heart and is responsible for cell-to-cell communication for electrical and physical purposes [108]. A combination of the reduced CX43 expression and sodium channel dysfunction causes a reduction in intercellular coupling current and membrane excitability [109], thus causing the prolongation of the QT interval and arrhythmias [94]. A prolonged QT interval can induce Torsade de Pointes of ventricular tachycardia or ventricular fibrillation, and sudden cardiac death [94,104]. Besides those factors, an increase of collagen content during fibrosis in the myocardium can disrupt the electrical conduction and arrhythmias [94]. The myocardium’s extracellular collagen matrix preserves the muscle fibers, cardiac cell alignment, and ventricular shape [110]. Cardiac remodeling to increase the collagen content causes the thickening of existing fibrillar collagen, stiff myocardium, and dysfunction in the left ventricular diastolic, a condition that is often seen in cardiac hypertrophy [110,111]. This reduced collagen content will also cause ventricular remodeling in HF [112]. Coronary blockage (atherosclerotic plaques) can also induce the breakdown of fibrillar collagen together with infarction (tissue death or necrosis) and loss of cardiac tissue integrity [94]. Thinning of these infarcted walls and ventricular dilation increases the risk of myocardial rupture, as seen in myocardial infarction (MI) patients [94].
4. Long Non-Coding RNAs in CVD
4.1. LncRNA in Atherosclerosis
In atherogenesis plaque development, endothelial cell (EC) dysfunction, inflammatory leukocytes with the release of cytokines and chemokines cause phenotypic changes in the vascular smooth muscle cells (VSMC) from a contractile-quiescent state to an activated proliferative state and migration with the subsequent formation of the extracellular matrix or the plaque [100,101]. EC dysfunction is an early event of atherosclerosis, and a few lncRNAs are involved in the regulation of EC function and structure (Table 3 and Figure 2) [113]. One such is the MALAT1 lncRNA, known as the EC pro-inflammatory and angiogenesis lncRNA [114]. Expression of MALAT1 is higher due to hyperglycemia, hypoxia, and oxidative stress [114]. Inhibition of MALAT1 expression causes the reduction of S-phase cyclins (CCNB1, CCNB2, and CCNA2), as well as the increase in expression of p21 and p27Kip1 (cell cycle inhibitors), resulting in suppression of EC proliferation during hypoxia [114]. Like MALAT1, the expression of lncRNA HOTTIP increases in the coronary artery tissue of CAD patients and EC-stimulated with an inflammatory cytokine, TNF-α [115]. The increased expression of this lncRNA HOTTIP promotes EC proliferation and migration via the Wnt/β-catenin pathway [115]. Another study found that lncRNA LINC00968 expression was higher in the coronary artery tissue of CAD patients. Upon stimulation of the oxidized low-density lipoprotein (oxLDL), this lncRNA expression was increased with EC proliferation and migration [116].

Table 3.
Summary of the reported long noncoding RNAs (lncRNAs) in cardiovascular disease (CVD). List of previously published lncRNAs in cardiovascular diseases and their information.
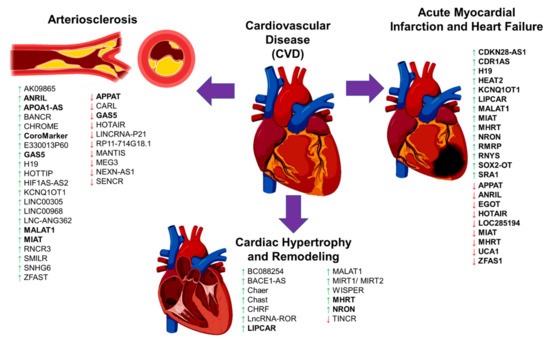
Figure 2.
The schematic diagram illustrates the long noncoding RNA (lncRNA) involvement in cardiovascular disease (CVD) complications. The diagram shows the involvement of previously published long noncoding RNAs (lncRNAs) in cardiovascular disease (CVD), focusing on arteriosclerosis, cardiac remodeling/hypertrophy, myocardial infarction, and heart failure. A green upward arrow indicates the upregulation of the lncRNA expression, a red downward arrow indicates the reduced expression, and the bold lncRNAs exist in the circulating biological samples for diagnostic potential.
LncRNAs are also involved in recruiting leukocytes and macrophage-derived foam cells, although the evidence is limited (Table 3 and Figure 2). One such example is the lncRNA RP5-833A20.1 that is increased in foam cells [130]. Overexpression of this lncRNA in macrophages caused the increase in inflammatory cytokines (IL-1β, IL-6, and TNFα) by inducing miR-382-5p expression leading to the reduced expression of nuclear factor I A (NFIA) [130]. Similarly, another study found that lncRNA E330013P60 expression increased macrophages isolated from diabetic mice [121]. The increase in this lncRNA expression caused a shift to inflammatory phenotypes in those macrophages and foam cell formation [121].
The expressions of three lncRNAs, GAS5, SNHG6, and ZFAS1, were high in the atherosclerotic plaques [122]. Overexpression of GAS5 in macrophage-derived foam cells increased the intracellular lipid accumulation, whereas the inhibition of this lncRNA expression inhibited intracellular lipid accumulation and its progression atherosclerosis [123]. The action of GAS5 lncRNA is mediated by its interaction with zeste homolog 2 (EZH2) to reduce the ATP-binding cassette transporter A1 (ABCA1) expression [123]. Interestingly, exosomes from the lncRNA GAS5-knockout macrophage cells prevented endothelial cell apoptosis upon the stimulation of oxidized LDL [155]. This evidence suggests that EC dysfunction could be prevented in the absence of GAS5. Another lncRNA associated with the atherosclerosis plaque is the MI-associated transcript (MIAT) that was upregulated in the serum samples of stroke patients with atherosclerosis, and in the mouse model of atherosclerosis [127,128]. LncRNA MIAT sponges the miR-181-b action to increase the STAT3 expression thus promotes the proliferation of VSMC [128]. Similarly, the lncRNA Cholesterol Homeostasis Regulator of MiRNA Expression (CHROME) was also upregulated in plasma patients of CAD and inflammatory cells in the atherosclerosis plaque [120]. This lncRNA CHROME interacts with miR-27b, miR-33a, miR-33b and miR-128 to regulate cellular cholesterol homeostasis [120].
Previous GWAS studies to determine the genetic susceptibility in CAD found that lncRNA ANRIL was strongly associated with a greater risk of having CAD [40]. Expression of this lncRNA ANRIL was higher in atherosclerotic plaques and peripheral blood mononuclear cells from CAD patients [118]. LncRNA ANRIL binds to the components of the polycomb repression complex-1 (PRC-1) and -2 (PRC-2) [156,157], thus initiating the suppression of the INK4 locus to decrease the expression of p15 and p16 proteins. Both p15 and p16 proteins are the controllers of cell division that limit the cell lifespan, and loss of these proteins will allow for aberrant cell proliferation as seen in VSMC and EC [158]. Other lncRNAs that regulate and promote VSMCs proliferation and migration are the smooth muscle–induced lncRNA enhances replication (SMILR) [131], AK09865 [117], H19 [124], LINC00305 [125], LNC-ANG362 [126], retinal ncRNA3 (RNCR3) [129], and BANCR [119].
Unlike the above lncRNAs with the adverse effects, the MEG3 is a protective lncRNA for the heart. MEG3 lncRNA expression was lower in EC stimulated with high glucose in vitro and in vivo conditions [139]. Reduced expression of MEG3 caused severe EC dysfunction, with evidence of microvascular leakage and inflammation, via the activation of the PI3K/Akt signaling pathway [139]. The mechanism of how MEG3 lncRNA protects EC function is partially by sponging the miR-9 action to regulate the angiogenesis and proliferation processes [159]. Another protective lncRNA is the cardiac apoptosis-related lncRNA (CARL), which was highly expressed in myocardial EC and was reduced during the development of myocardial infarction (MI) and atherosclerosis [133]. CARL lncRNA acts as a miRNA sponge to miR-539 and thus inhibits the suppression of the PHB2 gene, responsible for the regulation of mitochondrial function and apoptosis [134]. Overexpression of CARL caused a reduction of the PHB2 and BAX expressions and increased anti-apoptotic BCL2 [133] to prevent the cell apoptosis during MI.
Furthermore, lncRNA MANTIS was also lower in EC from idiopathic pulmonary arterial hypertension (IPAH) patients and animal models [138]. Interestingly, lncRNA MANTIS interacts with the Brg1 complex. This Brg1 protein complex is activated following the cardiac stress during cardiac remodeling and regulates the key genes in angiogenesis genes such as SOX18, SMAD6, and COUP-TFII [138]. Other lncRNAs that prevents VSMC proliferation and migration are the LncRNA-RP11-714G18.1 [137], NEXN antisense RNA 1 (NEXN-AS1) [140], smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA (SENCR) [141,160], HOX transcript antisense RNA (HOTAIR) [135], LINCRNA-P21 [136], and atherosclerotic plaque pathogenesis associated transcript (APPAT) [132]. Some of these lncRNAs have good diagnostic potential for CVD event prediction and will be discussed in the next section.
4.2. LncRNA in Heart Development
As for cardiac development, Braveheart (BVHT) lncRNA was the first lncRNA found to be necessary for the activation of important cardiac genes and transcription factors for cardiogenic differentiation and development [161,162]. Following that, many studies have identified other lncRNAs such as Fendrr [163], Linc1405 [164], HoxBlinc RNA [165], Handsdown (Hdn) [166] in mice, and (CAR)diac (M)esoderm (E)nhancer-associated (N)on-coding RNA (CARMEN) [167], Heart Brake LncRNA 1 (HBL1) [168] and Ppp1r1b [169] in human. The majority of these lncRNAs interact with Polycomb repressive complex-2 (PRC2), a histone methyltransferase responsible for epigenetic silencing during the development of the heart and body wall [170]. It is important to note that the exact mechanisms of how these lncRNAs regulate heart differentiation and development are not fully understood.
4.3. LncRNA in Cardiac Remodeling
Thickening of the ventricle wall and a reduction in the contractility of the heart are often seen in cardiac hypertrophy and remodeling, which could be fatal if the situation is prolonged. Among the lncRNAs that are involved in cardiac hypertrophy is the lncRNA cardiac hypertrophy-associated (Chast), which was increased in the mouse model of cardiac stress (transverse aortic constriction, TAC), as well as in human patients with aortic stenosis [145]. Overexpression of lncRNA Chast induced cardiac hypertrophy and remodeling in cardiomyocytes in vivo and in vitro models [145], whereas the silencing of this lncRNA prevented cardiac remodeling despite a stress-induction [145]. This cardiac hypertrophy action of Chast was shown due to its suppression of Pleckstrin homology domain-containing protein family M member 1 (Plekhml) expression that is responsible for cardiac autophagy [145]. Like Chast, the expression of cardiac-hypertrophy-associated epigenetic regulator (Chaer) lncRNA is increased during cardiac hypertrophy, and a heart-specific lncRNA [144]. Interestingly, a mouse model of lncRNA Chaer expression knockdown did not develop cardiac hypertrophy, with no substantial effect on the normal heart function and histology [144]. Therefore, this lncRNA Chaer is only involved in a pressure/stress-induced cardiac hypertrophy, and loss of its expression will not disrupt the normal heart function, hence this lncRNA is a good target for therapeutic strategy.
Another lncRNA is the cardiac hypertrophy-related factor (CHRF), which was highly expressed in a cellular model of cardiac hypertrophy [146]. Following the angiotensin II treatment (Ang-II), the expression of lncRNA CHRF was higher and this lncRNA suppressed the miR-489 expression as its endogenous microRNA sponge [146]. The reduction of miR-489 expression caused an increase of the myeloid differentiation primary response gene 88 (Myd88), thus initiating the cardiac hypertrophy [146]. LncRNA BC088254 expression was increased in the rat model of cardiac hypertrophy [142]. In this study [142], the expression of BC088254 was negatively correlated with the PHB2 gene, which is responsible for regulating mitochondrial function and apoptosis. Another reported lncRNA is the lncRNA-ROR, which was higher in cardiomyocytes of the TAC mouse model [147]. The mechanism of how lncRNA-ROR promotes cardiac hypertrophy is partially via its interaction with miR-133, in which they have an inverse relationship [147]. Loss of miR-133 expression led to the increase of cardiac myocytes and cardiac hypertrophy markers [171]. However, whether the inverse relationship between miR-133 and lncRNA-ROR expressions is due to microRNA sponge action or not is unknown. MALAT1 lncRNA is a well-known pro-inflammatory lncRNA, and its expression was higher in the rat model of myocardial ischemia-reperfusion (I/R) injury [148]. Inhibition of MALAT1 expression rescued the rats’ cardiac function due to a restoration of the PI3K/AKT signaling pathway [148]. Studies in the rat model of hypertension showed that MALAT1 expression was increased in the heart [149,150]. Overexpression of MALAT1 caused severe cardiac fibrosis and remodeling by reducing the expression of MyoD that is responsible for the contractile phenotype of the heart [149]. MALAT1 recruits the Suv39h1, a histone methyltransferase enzyme, to MyoD loci and causes H3K9me3 trimethylation to silence MyoD expression [149].
In terms of cardiac fibrosis, lncRNA WIsp2 SuPer-Enhancer associated RNA (WISPER) was reported to regulate fibrosis [153]. This lncRNA WISPER is a heart-enriched lncRNA, and its high expression was observed in MI-induced fibrosis of the mouse model and aortic stenosis patients [153]. Inhibition of this lncRNA expression prevented the MI-induced fibrosis and cardiac remodeling [153]. Besides that, in the mouse model of MI, Myocardial Infarction-Associated Transcript 1 (MIRT1) and 2 (MIRT2) expressions were significantly upregulated and correlated with the left ventricle remodeling [172]. However, the exact mechanism of how these lncRNAs promotes cardiac hypertrophy is unknown. In heart failure (HF) patients, antisense transcript of β-secretase-1 (BACE1), also known as lncRNA BACE1-AS1, was upregulated in left ventricle biopsies [143]. This high expression of BACE-AS1 increased EC apoptosis and silencing its expression removed this effect [143]. Another study of HF patients showed that the expression of a non-coding repressor of NFAT (NRON) lncRNA was higher in plasma samples of HF patients [152].
As for protective lncRNA in cardiac remodeling, lncRNA MHRT, known as the myosin heavy-chain associated RNA transcript, is the main lncRNA with this function. This lncRNA MHRT is abundantly and specifically expressed in the heart [151]. In the mouse model of transverse aortic constriction (TAC) (to induce cardiac stress), the expression of lncRNA MHRT was low in cardiac tissues, and this inhibition was due to Brg1-Hdac-Parp chromatin repressor complex-3 that was activated following cardiac stress [151]. The suppression of MHRT expression allows for cardiac remodeling to occur as the Brg1 protein has a specific dual-binding helicase site that can be occupied by MHRT or its target genomic DNA region. Thus, restoration of MHRT expression prevented the Brg1 action by binding to this helicase domain and sequestering Brg1 to prevent cardiac remodeling [151]. This MRHT-Brg1 negative feedback action may explain how the heart responds to injury in the T2D environment.
In contrast, plasma expression of MHRT was increased in HF patients [152]. The discrepancies in the findings of MHRT expression may be due to different CVD models and sampling, as one study used the heart tissues while the latter used plasma samples. However, this difference may also due to the feedback regulatory function of lncRNA MHRT to protect the heart. A previous study of rat cardiomyocytes in an in vitro acute MI model showed that in response to oxidative stress, lncRNA MHRT expression was upregulated to reduce cell apoptosis and released into the circulation [173]. Considering its important function, lncRNA MHRT may serve as an excellent potential biomarker for MI and HF. Similar to MHRT, lncRNA TINCR expression was lower in the TAC mouse model [154]. This lncRNA prevented cardiac hypertrophy by binding to the PRC2 complex and recruiting it to the CaMKII gene promoter. Silencing of the CaMKII gene attenuated Ang-II-induced cardiomyocyte hypertrophy [154]. Identification of these lncRNAs in CVD showed that most of these lncRNAs are heart-specific and may be potentially used as biomarkers for CVD and heart dysfunction.
5. Circulating LncRNA as Biomarkers for CVD
Multiple plasma proteins or peptides have been used as biomarkers to detect CVD events or predict their presence (Table 4). For example, to diagnose heart failure (HF), the measurement of brain natriuretic peptide (BNP) or N-terminal of prohormone BNP (NT-proBNP), ST-2, and atrial natriuretic peptide (ANP) are widely used [174]. For atherosclerosis or CAD, the level of troponin T (TnT), high sensitivity Troponin (HsTnT), and creatinine phosphokinase-MB are used [174,175]. However, the lack of early biomarkers to predict these CVD events (at least 1–2 years beforehand) is devastating, as early detection could help reduce the mortality risk. The presence of silent MI has a similar risk to the clinically diagnosed MI towards the HF incidence and death [176]. Therefore, prompting the urgent need to find more reliable biomarkers. One suggestion is to use the noncoding RNA biomarkers in the blood to improve the detection. A systematic review of circulating lncRNAs as a predictor of CVD future events showed that among the 30 studies, the lncRNA expression signatures have a moderate sensitivity with high specificity [177]. However, these lncRNAs have a strong potential to differentiate CVD patients from those without CVD or healthy controls (AUC: 0.85) [177].

Table 4.
Summary of the reported circulating long noncoding RNAs (lncRNAs) as biomarkers for cardiovascular disease. List of previously published circulating lncRNAs in cardiovascular diseases and their information.
Among those lncRNAs discussed in the previous section, some of them have been validated or investigated as potential biomarkers of HF for diagnostic purposes, especially in circulating blood or plasma samples. Examples are the increased expression of lncRNA MIAT [127,128], CHROME [120], NRON, and MHRT [152], and reduced expression of APPAT [132]. The first study to use lncRNA expression to predict CVD events is for detecting the left ventricular remodeling (LVM) after myocardial infarction (MI) in 788 patients [185]. In this study [185], a reduced expression of lncRNA uc022bqs.1 or LIPCAR was observed in the early stage of MI after the LVM, but its expression was increased in later stages as the condition worsened. Higher expression of LIPCAR was confirmed in the HF group with LVM, and even after the adjustment of other risk factors, the high expression of LIPCAR is an independent predictor of 3-year cardiovascular mortality (OR: 4.16) [185]. Importantly, for patients with the highest expression of LIPCAR, the mortality risk is even higher (OR: 32.58) [185]. Similarly, another study showed that expression of plasma LIPCAR correlated with the severity of coronary stenosis (AUC = 0.782) in the ST-segment elevation MI (STEMI) patients [186] and strongly predicts the CAD event (AUC: 0.722) [182].
In another study that involved HF patients, the levels of two lncRNAs, NRON and MHRT, in plasma samples could predict acute MI events with a sensitivity and specificity of 86.5% and 70.2%, respectively [152]. The expression of plasma MHRT was confirmed again in another study of chronic HF patients, in which the reduced expression of MHRT was associated with the worst outcomes of survival after the treatment [189]. A panel of nine lncRNAs (CDKN2B-AS1, EGOT, H19, HOTAIR, LOC285194, RMRP, RNY5, SOX2-OT, and SRA1) was able to differentiate the HF patients compared to controls [188]. In this study [188], CDKN2B-AS1, H19, RMRP, RNY5, SOX2-OT and SRA1 were upregulated, whereas EGOT, HOTAIR, and LOC285194 expressions were downregulated. High expression of lncRNA Heat2 was observed in peripheral blood mononuclear cells (PBMC) samples of HF patients, and this lncRNA expression is enriched in circulating immune cells [183]. Functional characterization of lncRNA Heat2 showed that this lncRNA regulates immune cell division, invasion, transmigration, and adhesion to EC [183].
As for predicting acute MI (AMI) alone, peripheral blood mononuclear cells (PBMC) derived lncRNA H19, MIAT, and MALAT1 were observed to be higher in AMI patients [181]. In this study, the diagnostic potential of lncRNA H19 to predict AMI was significant (AUC, 0.753; 95% CI, 0.689~0.817) and is associated with other CVD risk factors [181]. Another study showed that the plasma expression of lncRNA urothelial carcinoma-associated 1 (UCA1) was reduced in AMI 48 h after the event and started to increase afterward [190]. However, the diagnostic potential of this lncRNA UCA1 to predict AMI is only minimal (AUC: 0.757) [190]. One study showed that the expression of three lncRNAs, the potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1 (KCNQ1OT1), hypoxia-inducible factor 1A antisense RNA 2 (HIF1A-AS2) and MALAT1 were increased in PBMC samples in MI patients, whereas lncRNA ANRIL expression was reduced [184]. Comparison between STEMI and non-STEMI patients showed that ANRIL, KCNQ1OT1, MALAT1, and MIAT expression were lowered in those with STEMI [184]. Interestingly, the expression of both ANRIL and KCNQ1OT1 lncRNAs can correctly re-group the patients (left ventricular dysfunction) that was missed by the multiple clinical variables panel [184]. In contrast to the other lncRNAs, the expression of lncRNA APPAT was lower in blood samples of MI or angina pectoris patients with the potential to predict these CVD events at 78.72 % sensitivity and 93.02% specificity [132]. Similar to APPAT, lncRNA Zinc finger antisense 1 (ZFAS1) expression in plasma of AMI patients was reduced, however the diagnostic potential of this lncRNA to differentiate AMI from non-AMI patients, is minimal (AUC: 0.664) [179]. Interestingly, in this study [179], the expression of ZFAS1 was opposite to another lncRNA, Cdr1 antisense (CDR1AS), in which its expression was increased in AMI patients with a similar diagnostic potential (AUC: 0.671). As for detecting hypertension or cardiac hypertrophy, expression of three lncRNAs, MHRT, FENDRR and CARMEN was higher in PBMC samples of hypertensive patients, though no diagnostic potential was measured in this study [191].
In ischaemic stroke (IS) patients, the plasma expression of lncRNA MIAT expression was higher and correlated with the level of high-sensitivity C-reactive protein, infarct volume, and poor prognosis [187]. Moreover, this lncRNA expression in plasma could differentiate IS patients from the controls (AUC: 0.842) [187]. Another study also confirmed that lncRNA MIAT expression was higher in serum samples of the stroke patient with evidence of atherosclerotic plaques [127].
For predicting CAD, one study found that the higher plasma expression of lncRNA AC100865.1 or known as the CoroMarker, predicted the CAD event in a cohort of 221 CAD patients and 187 controls, with a sensitivity of 78.05% and a specificity of 86.49% [180]. Importantly, high expression of CoroMarker lncRNA was only evident in CAD patients, not with other CVD or metabolic diseases [180]. Another study also discovered three lncRNAs in peripheral blood mononuclear cells (PBMC) samples of CAD patients, in which the KCNQ1OT1, HIF1A-AS2, and apolipoprotein A-1 antisense RNA (APOA1-AS) lncRNAs were higher in CAD patients [178]. The diagnostic potential of each lncRNAs (area under ROC curve) was 0.865 (KCNQ1OT1), 0.852 (HIF1A-AS2), and 0.967 (APOA1-AS). The combination of these lncRNAs in a panel predicted the CAD event with a better AUC value of 0.990, and these lncRNA expressions also correlated with the levels of NT-proBNP and HsTnT markers [178]. Expression of H19 lncRNA was higher in the plasma of CAD patients, but this high expression of H19 only minimally predicted the CAD event (AUC: 0.631) [182]. In contrast, the expression of lncRNA GAS5 in the plasma samples of CAD patients was reduced and its diagnostic potential was shown to be high, with an AUC of 0.9783 [89], thus suggesting that GAS5 lncRNA is a very promising biomarker for CAD. Although these lncRNAs have been extensively analyzed to predict CVD events, their roles in CVD complications of T2D patients still require clarifications.
6. Circulating lncRNAs for CVD Complications in T2D
Previous findings of lncRNAs associated with T2D or CVD showed that some of these lncRNAs are overlapped between the two disease conditions (Figure 3). Thus, previous studies determined the diagnostic potential of these lncRNAs as biomarkers for CVD complications in T2D. One such lncRNA is the MALAT1, in which its expression was higher when cardiomyopathy occurred in diabetic rats [192]. A study of T2D patients showed that LINC-PINT expression in the plasma was lower in T2D patients. However, this reduction worsens when these T2D patients developed cardiomyopathy in the six-years follow-up visit [77]. Another is the lncRNA ANRIL expression, which was reported higher in T2D patients, and circulating ANRIL expression becomes much higher in T2D patients with MI, thus indicating the potential of ANRIL in detecting the CVD complication in T2D patients [92]. In another study of well-controlled diabetic patients, plasma expression of lncRNA LIPCAR correlated with left ventricular grade I diastolic dysfunction. The expressions of MIAT and SENCR lncRNAs correlated with the left ventricular remodeling in these diabetic patients [193]. As for other lncRNAs in Figure 3, these lncRNAs were reported separately in T2D and CVD conditions, and thus require validation in T2D subjects that developed CVD complications compared with those who did not. Despite being limited, this evidence suggests that circulating lncRNA expression could be used as the potential biomarkers to predict the early events of diabetic cardiomyopathy. Further identification of candidate lncRNAs is important as the T2D environment itself is a risk factor for CVD complications and may elevate the biomarkers’ levels more or differently. Nevertheless, these findings are still limited; thus, the interpretations of the results require more studies to confirm the conclusion.
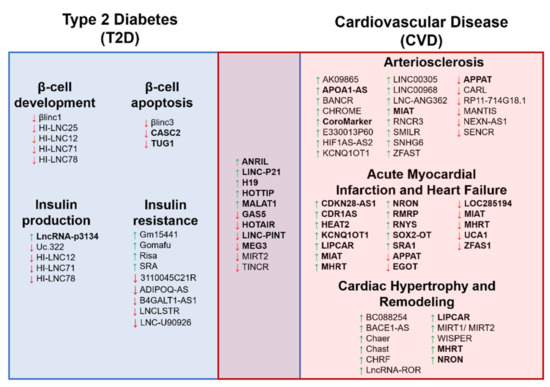
Figure 3.
The schematic diagram illustrates the long noncoding RNA (lncRNA) involvement in both Type 2 Diabetes (T2D) and cardiovascular disease (CVD) complications. The diagram shows the overlapping of the lncRNAs reported in both T2D and CVD conditions. A green upward arrow indicates the upregulation of the lncRNA expression, a red downward arrow indicates the reduced expression, and the bold lncRNAs exist in the circulating biological samples for diagnostic potential.
7. Conclusions
Despite the availability of various treatments for CVD and T2D, the mortality risk of T2D patients due to CVD remains high. Thus, identifying high-risk individuals among T2D patients is crucial for better disease management. One way of doing so is using the circulating long noncoding RNAs to complement the clinical variables to develop a better risk and prediction scoring system. Among the studies of lncRNAs as biomarkers, the most promising lncRNAs are from MI and HF of T2D studies, as the findings are more consistent and showed strong predictive values. Notably, most of these lncRNAs are heart-specific and exist in the circulating biological samples, making them potential biomarkers for future CVD events. Expressions of several lncRNAs (ANRIL, LINC-PINT, MALAT-1, LIPCAR, SENCR, and MIAT) have been measured in T2D individuals with CVD complications and could predict these CVD events. Although lncRNAs such as GAS5, H19, HOTAIR, HOTTIP, LINC-P21, MEG3, MIRT2, and TINCR, are reported in T2D and CVD individuals separately. Thus, further validation and confirmation of these lncRNAs in T2D individuals with CVD progression are needed. With more lncRNAs being identified and the growing understanding of the molecular mechanisms and roles of these lncRNAs, the application of using these lncRNAs as new biomarkers will allow for early identification of CVD complications in high-risk T2D patients.
Author Contributions
Conceptualization, S.A.S. and R.J.; writing—original draft preparation, N.I. and S.A.S.; writing—review and editing, N.A., N.A.A.M. and R.J.; visualization, N.I.; supervision, S.A.S., N.A., N.A.A.M. and R.J.; funding acquisition, S.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This manuscript was funded by the UKM University Research Grant, grant number “GUP-2018-069”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Some parts of the figures are made using the BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the review, in the data collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish.
References
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Strain, W.D.; Paldánius, P.M. Diabetes, cardiovascular disease and the microcirculation. Cardiovasc. Diabetol. 2018, 17, 57. [Google Scholar] [CrossRef] [PubMed]
- Gedebjerg, A.; Almdal, T.P.; Berencsi, K.; Rungby, J.; Nielsen, J.S.; Witte, D.R.; Friborg, S.; Brandslund, I.; Vaag, A.; Beck-Nielsen, H.; et al. Prevalence of micro- and macrovascular diabetes complications at time of type 2 diabetes diagnosis and associated clinical characteristics: A cross-sectional baseline study of 6958 patients in the Danish DD2 cohort. J. Diabetes Complicat. 2018, 32, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Henning, R.J. Type-2 diabetes mellitus and cardiovascular disease. Future Cardiol. 2018, 14, 491–509. [Google Scholar] [CrossRef] [PubMed]
- Rawshani, A.; Rawshani, A.; Franzén, S.; Eliasson, B.; Svensson, A.-M.; Miftaraj, M.; McGuire, D.K.; Sattar, N.; Rosengren, A.; Gudbjörnsdottir, S. Mortality and Cardiovascular Disease in Type 1 and Type 2 Diabetes. N. Engl. J. Med. 2017, 376, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Vassy, J.L.; Ho, Y.-L.; Song, R.J.; Gagnon, D.R.; Cho, K.; Wilson, P.W.F.; Phillips, L.S. Diabetes Mellitus-Related All-Cause and Cardiovascular Mortality in a National Cohort of Adults. J. Am. Heart Assoc. 2019, 8, e011295. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.J.; Imperatore, G.; Geiss, L.S.; Saydah, S.H.; Albright, A.L.; Ali, M.K.; Gregg, E.W. Trends and Disparities in Cardiovascular Mortality Among U.S. Adults With and Without Self-Reported Diabetes, 1988–2015. Diabetes Care 2018, 41, 2306–2315. [Google Scholar] [CrossRef] [PubMed]
- Anjana, R.M.; Mohan, V.; Rangarajan, S.; Gerstein, H.C.; Venkatesan, U.; Sheridan, P.; Dagenais, G.R.; Lear, S.A.; Teo, K.; Karsidag, K.; et al. Contrasting Associations Between Diabetes and Cardiovascular Mortality Rates in Low-, Middle-, and High-Income Countries: Cohort Study Data From 143,567 Individuals in 21 Countries in the PURE Study. Diabetes Care 2020, 43, 3094–3101. [Google Scholar] [CrossRef]
- Borhanuddin, B.; Mohd Nawi, A.; Shah, S.A.; Abdullah, N.; Syed Zakaria, S.Z.; Kamaruddin, M.A.; Velu, C.S.; Ismail, N.; Abdullah, M.S.; Ahmad Kamat, S.; et al. 10-Year Cardiovascular Disease Risk Estimation Based on Lipid Profile-Based and BMI-Based Framingham Risk ScoRes. across Multiple Sociodemographic Characteristics: The Malaysian Cohort Project. Sci. World J. 2018, 2018, 2979206. [Google Scholar] [CrossRef]
- De Rosa, S.; Arcidiacono, B.; Chiefari, E.; Brunetti, A.; Indolfi, C.; Foti, D.P. Type 2 Diabetes Mellitus and Cardiovascular Disease: Genetic and Epigenetic Links. Front. Endocrinol. 2018, 9, 2. [Google Scholar] [CrossRef]
- International Human Genome Sequencing, C. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Pertea, M. The human transcriptome: An unfinished story. Genes (Basel) 2012, 3, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Gen. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Gen. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Wendt, A.; Esguerra, J.L.S.; Eliasson, L. Islet microRNAs in health and type-2 diabetes. Curr. Opin. Pharm. 2018, 43, 46–52. [Google Scholar] [CrossRef]
- Wang, D.; Atanasov, A.G. The microRNAs Regulating Vascular Smooth Muscle Cell Proliferation: A Minireview. Int. J. Mol. Sci. 2019, 20, 324. [Google Scholar] [CrossRef]
- Das, A.; Samidurai, A.; Salloum, F.N. Deciphering Non-coding RNAs in Cardiovascular Health and Disease. Front. Cardiovasc. Med. 2018, 5, 73. [Google Scholar] [CrossRef]
- Chabior, A.; Pordzik, J.; Mirowska-Guzel, D.; Postuła, M. The role of acetylsalicylic acid and circulating microRNAs in primary prevention of cardiovascular events in patients with Diabetes Mellitus Type 2—A Review. Ann. Agric. Environ. Med. 2019, 26, 512–522. [Google Scholar] [CrossRef]
- Regazzi, R. MicroRNAs as therapeutic targets for the treatment of diabetes mellitus and its complications. Expert Opin. Targets 2018, 22, 153–160. [Google Scholar] [CrossRef]
- Skuratovskaia, D.; Vulf, M.; Komar, A.; Kirienkova, E.; Litvinova, L. Promising Directions in Atherosclerosis Treatment Based on Epigenetic Regulation Using MicroRNAs and Long Noncoding RNAs. Biomolecules 2019, 9, 226. [Google Scholar] [CrossRef]
- He, X.; Ou, C.; Xiao, Y.; Han, Q.; Li, H.; Zhou, S. LncRNAs: Key players and novel insights into diabetes mellitus. Oncotarg 2017, 8, 71325–71341. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Somoza, A.S.; Devaux, Y.; Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxid. Redox Signal. 2018, 29, 880–901. [Google Scholar] [CrossRef] [PubMed]
- Hermans-Beijnsberger, S.; van Bilsen, M.; Schroen, B. Long non-coding RNAs in the failing heart and vasculature. Noncoding Rna Res. 2018, 3, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Zaiou, M. circRNAs Signature as Potential Diagnostic and Prognostic Biomarker for Diabetes Mellitus and Related Cardiovascular Complications. Cells 2020, 9, 659. [Google Scholar] [CrossRef]
- Gurha, P. Noncoding RNAs in cardiovascular diseases. Curr. Opin. Cardiol. 2019, 34, 241–245. [Google Scholar] [CrossRef]
- Li, M.; Duan, L.; Li, Y.; Liu, B. Long noncoding RNA/circular noncoding RNA–miRNA–mRNA axes in cardiovascular diseases. Life Sci. 2019, 233, 116440. [Google Scholar] [CrossRef]
- Jusic, A.; Devaux, Y. Mitochondrial noncoding RNA-regulatory network in cardiovascular disease. Basic Res. Cardiol. 2020, 115, 23. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, J.; Du, X.; Fu, X. The interplay between noncoding RNAs and insulin in diabetes. Cancer Lett. 2018, 419, 53–63. [Google Scholar] [CrossRef]
- Loganathan, T.S.; Sulaiman, S.A.; Abdul Murad, N.A.; Shah, S.A.; Abdul Gafor, A.H.; Jamal, R.; Abdullah, N. Interactions Among Non-Coding RNAs in Diabetic Nephropathy. Front. Pharm. 2020, 11, 191. [Google Scholar] [CrossRef]
- Sulaiman, S.A.; Muhsin, N.I.A.; Jamal, R. Regulatory Non-coding RNAs Network in Non-alcoholic Fatty Liver Disease. Front. Physiol. 2019, 10, 279. [Google Scholar] [CrossRef]
- Shi, Q.; Yang, X. Circulating MicroRNA and Long Noncoding RNA as Biomarkers of Cardiovascular Diseases. J. Cell Physiol. 2016, 231, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Shields, E.J.; Petracovici, A.F.; Bonasio, R. lncRedibly versatile: Biochemical and biological functions of long noncoding RNAs. Biochem. J. 2019, 476, 1083–1104. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018, 60, 25. [Google Scholar] [CrossRef]
- Tavares, R.C.A.; Pyle, A.M.; Somarowthu, S. Phylogenetic Analysis with Improved Parameters Reveals Conservation in lncRNA Structures. J. Mol. Biol. 2019, 431, 1592–1603. [Google Scholar] [CrossRef]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef]
- Tsai, F.-J.; Yang, C.-F.; Chen, C.-C.; Chuang, L.-M.; Lu, C.-H.; Chang, C.-T.; Wang, T.-Y.; Chen, R.-H.; Shiu, C.-F.; Liu, Y.-M.; et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Gen. 2010, 6, e1000847. [Google Scholar] [CrossRef]
- Holdt, L.M.; Teupser, D. Long Noncoding RNA ANRIL: Lnc-ing Genetic Variation at the Chromosome 9p21 Locus to Molecular Mechanisms of Atherosclerosis. Front. Cardiovasc. Med. 2018, 5, 145. [Google Scholar] [CrossRef]
- Morán, I.; Akerman, I.; van de Bunt, M.; Xie, R.; Benazra, M.; Nammo, T.; Arnes, L.; Nakić, N.; García-Hurtado, J.; Rodríguez-Seguí, S.; et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012, 16, 435–448. [Google Scholar] [CrossRef]
- Cao, Z.; Yao, F.; Lang, Y.; Feng, X. Elevated Circulating LINC-P21 Serves as a Diagnostic Biomarker of Type 2 Diabetes Mellitus and Regulates Pancreatic β-cell Function by Sponging miR-766-3p to Upregulate NR3C2. Exp. Clin. Endocrinol. Diabetes 2020. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cell Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Arnes, L.; Akerman, I.; Balderes, D.A.; Ferrer, J.; Sussel, L. βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev. 2016, 30, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Motterle, A.; Gattesco, S.; Peyot, M.-L.; Esguerra, J.L.S.; Gomez-Ruiz, A.; Laybutt, D.R.; Gilon, P.; Burdet, F.; Ibberson, M.; Eliasson, L.; et al. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol. Metab. 2017, 6, 1407–1418. [Google Scholar] [CrossRef]
- Jin, F.; Wang, N.; Zhu, Y.; You, L.; Wang, L.; De, W.; Tang, W. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell Physiol. Biochem. 2017, 43, 2062–2073. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, J.; Xu, P.; Gui, R. Long Non-coding RNA GAS5 Maintains Insulin Secretion by Regulating Multiple miRNAs in INS-1 832/13 Cells. Front. Mol. Biosci. 2020, 7, 559267. [Google Scholar] [CrossRef]
- Alfaifi, M.; Verma, A.K.; Alshahrani, M.Y.; Joshi, P.C.; Alkhathami, A.G.; Ahmad, I.; Hakami, A.R.; Beg, M.M.A. Assessment of Cell-Free Long Non-Coding RNA-H19 and miRNA-29a, miRNA-29b Expression and Severity of Diabetes. Diabetes Metab. Syndr. Obes. 2020, 13, 3727–3737. [Google Scholar] [CrossRef]
- Akerman, I.; Tu, Z.; Beucher, A.; Rolando, D.M.Y.; Sauty-Colace, C.; Benazra, M.; Nakic, N.; Yang, J.; Wang, H.; Pasquali, L.; et al. Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab. 2017, 25, 400–411. [Google Scholar] [CrossRef]
- Xu, X.; Tian, J.; Li, Q.Y. Downregulation of HOTTIP regulates insulin secretion and cell cycle in islet β cells via inhibiting MEK/ERK pathway. Eur. Rev. Med. Pharm. Sci. 2018, 22, 4962–4968. [Google Scholar]
- Zhu, Y.; Li, Y.; Dai, C.; Sun, L.; You, L.; De, W.; Yuan, Q.; Wang, N.; Chen, Y. Inhibition of Lincpint expression affects insulin secretion and apoptosis in mouse pancreatic β cells. Int. J. Biochem. Cell Biol. 2018, 104, 171–179. [Google Scholar] [CrossRef]
- Kameswaran, V.; Bramswig, N.C.; McKenna, L.B.; Penn, M.; Schug, J.; Hand, N.J.; Chen, Y.; Choi, I.; Vourekas, A.; Won, K.J.; et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014, 19, 135–145. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Wang, N.; Yin, D.; Wang, L.; Jin, F.; Zhu, Y.; Yuan, Q.; De, W. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J. Cell Physiol. 2016, 231, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.D.; Zhang, E.B.; You, L.H.; Wang, N.; Wang, L.T.; Jin, F.Y.; Zhu, Y.N.; Cao, L.H.; Yuan, Q.X.; De, W.; et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell Physiol. Biochem. 2015, 35, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Rong, C.; Pan, F.; Xiang, L.; Wang, X.; Hu, Y. Expression characteristics of long noncoding RNA uc.322 and its effects on pancreatic islet function. J. Cell Biochem. 2018, 119, 9239–9248. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Li, J.; Feng, S.; Li, Y.; Tan, L. Long noncoding RNA Gomafu upregulates Foxo1 expression to promote hepatic insulin resistance by sponging miR-139-5p. Cell Death Dis. 2018, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, Y.; Wang, X.J.; Duan, B.H.; Li, L. HOTAIR participates in hepatic insulin resistance via regulating SIRT1. Eur. Rev. Med. Pharm. Sci. 2018, 22, 7883–7890. [Google Scholar]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wu, Y.B.; Zhou, J.; Kang, D.M. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem. Biophys. Res. Commun. 2016, 469, 319–325. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Sun, C.; Zhuo, S.; He, Z.; Wang, H.; Yan, M.; Liu, J.; Luan, Y.; Dai, C.; et al. Down-regulation of Risa improves insulin sensitivity by enhancing autophagy. FASEB J. 2016, 30, 3133–3145. [Google Scholar] [CrossRef]
- Chen, G.; Yu, D.; Nian, X.; Liu, J.; Koenig, R.J.; Xu, B.; Sheng, L. LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase (ATGL). Sci. Rep. 2016, 6, 35531. [Google Scholar] [CrossRef]
- Wang, J.; Yang, W.; Chen, Z.; Chen, J.; Meng, Y.; Feng, B.; Sun, L.; Dou, L.; Li, J.; Cui, Q.; et al. Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis. Diabetes 2018, 67, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Li, D.; Sun, H.; Li, M.; Hu, H. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene 2019, 700, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhou, Y.; Yuan, Q.; Gao, Y.; Wang, Y.; Wang, X.; Cui, X.; Xu, P.; Ji, C.; Guo, X.; et al. Dynamic transcriptome profile in db/db skeletal muscle reveal critical roles for long noncoding RNA regulator. Int. J. Biochem. Cell Biol. 2018, 104, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Gui, W.; Zhu, W.F.; Zhu, Y.; Tang, S.; Zheng, F.; Yin, X.; Lin, X.; Li, H. LncRNAH19 improves insulin resistance in skeletal muscle by regulating heterogeneous nuclear ribonucleoprotein A1. Cell Commun. Signal. 2020, 18, 173. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Liu, Y.; Xu, Y.; Jiang, Y.; Zhang, N.; Wang, Z.; Carmichael, G.G.; Taylor, H.S.; Li, D.; Huang, Y. H19 lncRNA Promotes Skeletal Muscle Insulin Sensitivity in Part by Targeting AMPK. Diabetes 2018, 67, 2183–2198. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.X.; Zou, Y.; Wang, G.T.; Huang, S.H.; Zhou, Y.J.; Zhou, Y.J. lnc TINCR induced by NOD1 mediates inflammatory response in 3T3-L1 adipocytes. Gene 2019, 698, 150–156. [Google Scholar] [CrossRef]
- Cai, R.; Sun, Y.; Qimuge, N.; Wang, G.; Wang, Y.; Chu, G.; Yu, T.; Yang, G.; Pang, W. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 420–432. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Lu, S.; Yin, L.; Zong, C.; Cui, S.; Qin, D.; Yang, Y.; Guan, Q.; Li, X.; et al. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int. J. Obes. (Lond.) 2017, 41, 299–308. [Google Scholar] [CrossRef]
- Arnes, L.; Sussel, L. Epigenetic modifications and long noncoding RNAs influence pancreas development and function. Trends Genet. 2015, 31, 290–299. [Google Scholar] [CrossRef]
- Christensen, A.A.; Gannon, M. The Beta Cell in Type 2 Diabetes. Curr. Diab. Rep. 2019, 19, 81. [Google Scholar] [CrossRef] [PubMed]
- Sussel, L.; Kalamaras, J.; Hartigan-O’Connor, D.J.; Meneses, J.J.; Pedersen, R.A.; Rubenstein, J.L.; German, M.S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development (Cambridge, English) 1998, 125, 2213–2221. [Google Scholar]
- Voight, B.F.; Scott, L.J.; Steinthorsdottir, V.; Morris, A.P.; Dina, C.; Welch, R.P.; Zeggini, E.; Huth, C.; Aulchenko, Y.S.; Thorleifsson, G.; et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010, 42, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Gaertner, B.; van Heesch, S.; Schneider-Lunitz, V.; Schulz, J.F.; Witte, F.; Blachut, S.; Nguyen, S.; Wong, R.; Matta, I.; Hübner, N.; et al. A human ESC-based screen identifies a role for the translated lncRNA LINC00261 in pancreatic endocrine differentiation. ELife 2020, 9, e58659. [Google Scholar] [CrossRef] [PubMed]
- Fadista, J.; Vikman, P.; Laakso, E.O.; Mollet, I.G.; Esguerra, J.L.; Taneera, J.; Storm, P.; Osmark, P.; Ladenvall, C.; Prasad, R.B.; et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. PNAS 2014, 111, 13924–13929. [Google Scholar] [CrossRef]
- Zha, T.; Su, F.; Liu, X.; Yang, C.; Liu, L. Role of Long Non-Coding RNA (LncRNA) LINC-PINT Downregulation in Cardiomyopathy and Retinopathy Progression Among Patients with Type 2 Diabetes. Med. Sci. Monit. 2019, 25, 8509–8514. [Google Scholar] [CrossRef]
- Prentki, M.; Nolan, C.J. Islet beta cell failure in type 2 diabetes. J. Clin. Invest. 2006, 116, 1802–1812. [Google Scholar] [CrossRef]
- Bagge, A.; Clausen, T.R.; Larsen, S.; Ladefoged, M.; Rosenstierne, M.W.; Larsen, L.; Vang, O.; Nielsen, J.H.; Dalgaard, L.T. MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochem. Biophys. Res. Commun. 2012, 426, 266–272. [Google Scholar] [CrossRef]
- Sanchez-Parra, C.; Jacovetti, C.; Dumortier, O.; Lee, K.; Peyot, M.L.; Guay, C.; Prentki, M.; Laybutt, D.R.; Van Obberghen, E.; Regazzi, R. Contribution of the Long Noncoding RNA H19 to β-Cell Mass Expansion in Neonatal and Adult Rodents. Diabetes 2018, 67, 2254–2267. [Google Scholar] [CrossRef]
- Pielok, A.; Marycz, K. Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance. Int. J. Mol. Sci. 2020, 21, 4182. [Google Scholar] [CrossRef]
- Carter, G.; Miladinovic, B.; Patel, A.A.; Deland, L.; Mastorides, S.; Patel, N.A. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015, 4, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zheng, J.; Hu, X.; Chen, L. Dysregulated expression of long noncoding RNAs serves as diagnostic biomarkers of type 2 diabetes mellitus. Endocrine 2019, 65, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Z.; Gao, C.; Rao, L.; Hao, P.; Jian, D.; Li, W.; Tang, H.; Li, M. The Diagnostic Value of Whole Blood lncRNA ENST00000550337.1 for Pre-Diabetes and Type 2 Diabetes Mellitus. Exp. Clin. Endocrinol. Diabetes 2017, 125, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Erfanian Omidvar, M.; Ghaedi, H.; Kazerouni, F.; Kalbasi, S.; Shanaki, M.; Miraalamy, G.; Zare, A.; Rahimipour, A. Clinical significance of long noncoding RNA VIM-AS1 and CTBP1-AS2 expression in type 2 diabetes. J. Cell Biochem. 2019, 120, 9315–9323. [Google Scholar] [CrossRef]
- Saeidi, L.; Ghaedi, H.; Sadatamini, M.; Vahabpour, R.; Rahimipour, A.; Shanaki, M.; Mansoori, Z.; Kazerouni, F. Long non-coding RNA LY86-AS1 and HCG27_201 expression in type 2 diabetes mellitus. Mol. Biol. Rep. 2018, 45, 2601–2608. [Google Scholar] [CrossRef]
- Mansoori, Z.; Ghaedi, H.; Sadatamini, M.; Vahabpour, R.; Rahimipour, A.; Shanaki, M.; Saeidi, L.; Kazerouni, F. Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol. Biol. Rep. 2018, 45, 1227–1233. [Google Scholar] [CrossRef]
- Wang, L.; Su, N.; Zhang, Y.; Wang, G. Clinical Significance of Serum lncRNA Cancer Susceptibility Candidate 2 (CASC2) for Chronic Renal Failure in Patients with Type 2 Diabetes. Med. Sci. Monit. 2018, 24, 6079–6084. [Google Scholar] [CrossRef]
- Yin, Q.; Wu, A.; Liu, M. Plasma Long Non-Coding RNA (lncRNA) GAS5 is a New Biomarker for Coronary Artery Disease. Med. Sci. Monit. 2017, 23, 6042–6048. [Google Scholar] [CrossRef]
- Kong, Y.; Sharma, R.B.; Nwosu, B.U.; Alonso, L.C. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia 2016, 59, 1579–1593. [Google Scholar] [CrossRef]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.M. Expression and function of lncRNA ANRIL in a mouse model of acute myocardial infarction combined with type 2 diabetes mellitus. J. Chin. Med. Assoc. 2019, 82, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-J.; Bai, T.; Zhi, L.-P.; Liu, Z.-H.; Liu, T.; Xue, H.; Yang, H.-H.; Yang, X.-H.; Zhang, M.; Niu, Y.-R.; et al. Analysis of long noncoding RNA-associated competing endogenous RNA network in glucagon-like peptide-1 receptor agonist-mediated protection in β cells. World J. Diabetes 2020, 11, 374–390. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, P.S.; Polegato, B.F.; Minicucci, M.F.; Paiva, S.A.R.; Zornoff, L.A.M. Cardiac Remodeling: Concepts, Clinical Impact, Pathophysiological Mechanisms and Pharmacologic Treatment. ARQ Bras. Cardiol. 2016, 106, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Niiranen, T.J.; Kalesan, B.; Mitchell, G.F.; Vasan, R.S. Relative Contributions of Pulse Pressure and Arterial Stiffness to Cardiovascular Disease. Hypertension 2019, 73, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Sharif, S.; Visseren, F.L.J.; Spiering, W.; de Jong, P.A.; Bots, M.L.; Westerink, J.; SMART study group. Arterial stiffness as a risk factor for cardiovascular events and all-cause mortality in people with Type 2 diabetes. Diabet. Med. 2019, 36, 1125–1132. [Google Scholar] [CrossRef]
- Gomez-Sanchez, L.; Garcia-Ortiz, L.; Patino-Alonso, M.C.; Recio-Rodriguez, J.I.; Feuerbach, N.; Marti, R.; Agudo-Conde, C.; Rodriguez-Sanchez, E.; Maderuelo-Fernandez, J.A.; Ramos, R.; et al. Glycemic markers and relation with arterial stiffness in Caucasian subjects of the MARK study. PLoS ONE 2017, 12, e0175982. [Google Scholar] [CrossRef]
- Rubin, J.; Nambi, V.; Chambless, L.E.; Steffes, M.W.; Juraschek, S.P.; Coresh, J.; Sharrett, A.R.; Selvin, E. Hyperglycemia and arterial stiffness: The Atherosclerosis Risk in the Communities study. Atherosclerosis 2012, 225, 246–251. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Shin, M.-H.; Choi, J.-S.; Rhee, J.-A.; Nam, H.-S.; Jeong, S.-K.; Park, K.-S.; Ryu, S.-Y.; Choi, S.-W.; Kim, B.-H.; et al. HbA1c is significantly associated with arterial stiffness but not with carotid atherosclerosis in a community-based population without type 2 diabetes: The Dong-gu study. Atherosclerosis 2016, 247, 1–6. [Google Scholar] [CrossRef]
- Rask-Madsen, C.; King, G.L. Vascular complications of diabetes: Mechanisms of injury and protective factors. Cell Metab. 2013, 17, 20–33. [Google Scholar] [CrossRef]
- Kim, J.K. Endothelial nuclear factor κB in obesity and aging. Circulation 2012, 125, 1081–1083. [Google Scholar] [CrossRef]
- Che Man, R.; Sulaiman, N.; Ishak, M.F.; Bt Hj Idrus, R.; Abdul Rahman, M.R.; Yazid, M.D. The Effects of Pro-Inflammatory and Anti-Inflammatory Agents for the Suppression of Intimal Hyperplasia: An Evidence-Based Review. Int. J. Environ. Res. Public Health 2020, 17, 7825. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Franquesa, A.; Patti, M.-E. Insulin Resistance and Mitochondrial Dysfunction. In Mitochondrial Dynamics in Cardiovascular Medicine; Santulli, G., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 465–520. [Google Scholar] [CrossRef]
- Heusch, G.; Libby, P.; Gersh, B.; Yellon, D.; Böhm, M.; Lopaschuk, G.; Opie, L. Cardiovascular remodelling in coronary artery disease and heart failure. Lancet 2014, 383, 1933–1943. [Google Scholar] [CrossRef]
- Seferović, P.M.; Paulus, W.J. Clinical diabetic cardiomyopathy: A two-faced disease with restrictive and dilated phenotypes. Eur. Heart J. 2015, 36, 1718–1727. [Google Scholar] [CrossRef]
- Coronel, R.; Wilders, R.; Verkerk, A.O.; Wiegerinck, R.F.; Benoist, D.; Bernus, O. Electrophysiological changes in heart failure and their implications for arrhythmogenesis. Biochim. Biophys. Acta 2013, 1832, 2432–2441. [Google Scholar] [CrossRef] [PubMed]
- Wiegerinck, R.F.; van Veen, T.A.B.; Belterman, C.N.; Schumacher, C.A.; Noorman, M.; de Bakker, J.M.T.; Coronel, R. Transmural dispersion of refractoriness and conduction velocity is associated with heterogeneously reduced connexin43 in a rabbit model of heart failure. Heart Rhythm. 2008, 5, 1178–1185. [Google Scholar] [CrossRef]
- Severs, N.J.; Bruce, A.F.; Dupont, E.; Rothery, S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res. 2008, 80, 9–19. [Google Scholar] [CrossRef]
- Wilders, R. Arrhythmogenic Right Ventricular Cardiomyopathy: Considerations from in Silico Experiments. Front. Physiol. 2012, 3, 168. [Google Scholar] [CrossRef]
- Tao, G.; Levay, A.K.; Peacock, J.D.; Huk, D.J.; Both, S.N.; Purcell, N.H.; Pinto, J.R.; Galantowicz, M.L.; Koch, M.; Lucchesi, P.A.; et al. Collagen XIV is important for growth and structural integrity of the myocardium. J. Mol. Cell Cardiol. 2012, 53, 626–638. [Google Scholar] [CrossRef]
- Brower, G.L.; Gardner, J.D.; Forman, M.F.; Murray, D.B.; Voloshenyuk, T.; Levick, S.P.; Janicki, J.S. The relationship between myocardial extracellular matrix remodeling and ventricular function. Eur. J. Cardiothorac. Surg 2006, 30, 604–610. [Google Scholar] [CrossRef]
- Janicki, J.S.; Brower, G.L. The role of myocardial fibrillar collagen in ventricular remodeling and function. J. Card. Fail. 2002, 8, S319–S325. [Google Scholar] [CrossRef]
- Haemmig, S.; Simion, V.; Yang, D.; Deng, Y.; Feinberg, M.W. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr. Opin. Cardiol. 2017, 32, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Chen, R.; Lin, F.; Mai, A.; Chen, J.; Li, H.; Xu, Z.; Dong, S. Long noncoding RNA HOTTIP promotes endothelial cell proliferation and migration via activation of the Wnt/β-catenin pathway. J. Cell. Biochem. 2018, 119, 2797–2805. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, Z.; Zhang, W.; Wang, Y. Long noncoding RNA LINC00968 promotes endothelial cell proliferation and migration via regulating miR-9-3p expression. J. Cell Biochem. 2019, 120, 8214–8221. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell-Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272. [Google Scholar] [CrossRef]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gäbel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL Expression Is Associated With Atherosclerosis Risk at Chromosome 9p21. Arter. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Zhang, L.; Li, X. LncRNA BANCR facilitates vascular smooth muscle cell proliferation and migration through JNK pathway. Oncotarg 2017, 8, 114568–114575. [Google Scholar] [CrossRef]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Reddy, M.A.; Chen, Z.; Park, J.T.; Wang, M.; Lanting, L.; Zhang, Q.; Bhatt, K.; Leung, A.; Wu, X.; Putta, S.; et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 2014, 63, 4249–4261. [Google Scholar] [CrossRef]
- Chen, L.; Yao, H.; Hui, J.-Y.; Ding, S.-H.; Fan, Y.-L.; Pan, Y.-H.; Chen, K.-H.; Wan, J.-Q.; Jiang, J.-Y. Global transcriptomic study of atherosclerosis development in rats. Gene 2016, 592, 43–48. [Google Scholar] [CrossRef]
- Meng, X.-D.; Yao, H.-H.; Wang, L.-M.; Yu, M.; Shi, S.; Yuan, Z.-X.; Liu, J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE-/- Mice. Mol. Nucleic Acids 2020, 19, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, L.; Mao, Y.; Nan, G. Long Noncoding RNA-H19 Contributes to Atherosclerosis and Induces Ischemic Stroke via the Upregulation of Acid Phosphatase 5. Front. Neurol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-D.; Wang, W.-T.; Xiong, J.; Xie, X.-M.; Cui, S.-S.; Zhao, Z.-G.; Li, M.J.; Zhang, Z.-Q.; Hao, D.-L.; Zhao, X.; et al. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NF-κB pathway in human monocytes. Sci. Rep. 2017, 7, 46204. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Trac, C.; Jin, W.; Lanting, L.; Akbany, A.; Sætrom, P.; Schones, D.E.; Natarajan, R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ. Res. 2013, 113, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-M.; Yang, S.; Xia, Y.-P.; Hu, R.-T.; Chen, S.; Li, B.-W.; Chen, S.-L.; Luo, X.-Y.; Mao, L.; Li, Y.; et al. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis. 2019, 10, 138. [Google Scholar] [CrossRef]
- Zhong, X.; Ma, X.; Zhang, L.; Li, Y.; Li, Y.; He, R. MIAT promotes proliferation and hinders apoptosis by modulating miR-181b/STAT3 axis in ox-LDL-induced atherosclerosis cell models. Biomed. Pharm. 2018, 97, 1078–1085. [Google Scholar] [CrossRef]
- Shan, K.; Jiang, Q.; Wang, X.Q.; Wang, Y.N.Z.; Yang, H.; Yao, M.D.; Liu, C.; Li, X.M.; Yao, J.; Liu, B.; et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016, 7, e2248. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Zhao, J.-Y.; Li, S.-F.; Huang, J.-L.; Qiu, Y.-R.; Ma, X.; Wu, S.-G.; Chen, Z.-P.; Hu, Y.-R.; Yang, J.-Y.; et al. RP5-833A20.1/miR-382-5p/NFIA-Dependent Signal Transduction Pathway Contributes to the Regulation of Cholesterol Homeostasis and Inflammatory Reaction. Arter. Thromb. Vasc. Biol. 2015, 35, 87–101. [Google Scholar] [CrossRef]
- Ballantyne, M.D.; Pinel, K.; Dakin, R.; Vesey, A.T.; Diver, L.; Mackenzie, R.; Garcia, R.; Welsh, P.; Sattar, N.; Hamilton, G.; et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation 2016, 133, 2050–2065. [Google Scholar] [CrossRef]
- Meng, F.; Yan, J.; Ma, Q.; Jiao, Y.; Han, L.; Xu, J.; Yang, F.; Liu, J. Expression status and clinical significance of lncRNA APPAT in the progression of atherosclerosis. PeerJ 2018, 6, e4246. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.J.; Zhang, H.S. LncRNA-CARl in a rat model of myocardial infarction. Eur. Rev. Med. Pharm. Sci. 2018, 22, 4332–4340. [Google Scholar]
- Wang, K.; Long, B.; Zhou, L.-Y.; Liu, F.; Zhou, Q.-Y.; Liu, C.-Y.; Fan, Y.-Y.; Li, P.-F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014, 5, 3596. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Meng, K.; Jiang, L.; Zhong, Y.; Yang, Y.; Lan, Y.; Zeng, Q.; Cheng, L. Thymic stromal lymphopoietin-induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci. Rep. 2017, 37, BSR20170351. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cai, J.; Han, Y.; Chen, J.; Huang, Z.-P.; Chen, C.; Cai, Y.; Huang, H.; Yang, Y.; Liu, Y.; et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014, 130, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Xu, B.-M.; Tang, W.-H.; Ye, Z.-D.; Huang, C.; Ma, X.; Zhao, J.-J.; Guo, F.-X.; Kang, C.-M.; et al. LncRNA-RP11-714G18.1 suppresses vascular cell migration via directly targeting LRP2BP. Immunol. Cell Biol. 2018, 96, 175–189. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Günther, S.; et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef]
- Qiu, G.-Z.; Tian, W.; Fu, H.-T.; Li, C.-P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Guo, F.-X.; Xu, Y.-J.; Li, P.; Lu, Z.-F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.H.; Kang, C.-M.; et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J. Clin. Invest. 2019, 129, 1115–1128. [Google Scholar] [CrossRef]
- Boulberdaa, M.; Scott, E.; Ballantyne, M.; Garcia, R.; Descamps, B.; Angelini, G.D.; Brittan, M.; Hunter, A.; McBride, M.; McClure, J.; et al. A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells. Mol. Ther. 2016, 24, 978–990. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Liang, J. Unraveling the Expression Profiles of Long Noncoding RNAs in Rat Cardiac Hypertrophy and Functions of lncRNA BC088254 in Cardiac Hypertrophy Induced by Transverse Aortic Constriction. Cardiology 2016, 134, 84–98. [Google Scholar] [CrossRef]
- Greco, S.; Zaccagnini, G.; Fuschi, P.; Voellenkle, C.; Carrara, M.; Sadeghi, I.; Bearzi, C.; Maimone, B.; Castelvecchio, S.; Stellos, K.; et al. Increased BACE1-AS long noncoding RNA and β-amyloid levels in heart failure. Cardiovasc. Res. 2017, 113, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.-J.; Ji, Y.-X.; Zhang, P.; Deng, K.-Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8, ra322–ra326. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.-Y.; Long, B.; Yuan, S.-M.; Wang, Y.; Liu, C.-Y.; Sun, T.; Zhang, X.-J.; Li, P.-F. The Long Noncoding RNA CHRF Regulates Cardiac Hypertrophy by Targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhou, X.; Huang, J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS ONE 2016, 11, e0152767. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Cheng, Y.T.; Yan, L.X.; Krittanawong, C.; Qian, W.; Zhang, H.J. LncRNA MALAT1 knockdown alleviates myocardial apoptosis in rats with myocardial ischemia-reperfusion through activating PI3K/AKT signaling pathway. Eur. Rev. Med. Pharm. Sci. 2019, 23, 10523–10531. [Google Scholar]
- Li, D.; Zhang, C.; Li, J.; Che, J.; Yang, X.; Xian, Y.; Li, X.; Cao, C. Long non-coding RNA MALAT1 promotes cardiac remodeling in hypertensive rats by inhibiting the transcription of MyoD. Aging (Albany NY) 2019, 11, 8792–8809. [Google Scholar] [CrossRef]
- Xue, Y.-Z.; Li, Z.-J.; Liu, W.-T.; Shan, J.-J.; Wang, L.; Su, Q. Down-regulation of lncRNA MALAT1 alleviates vascular lesion and vascular remodeling of rats with hypertension. Aging (Albany NY) 2019, 11, 5192–5205. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef]
- Xuan, L.; Sun, L.; Zhang, Y.; Huang, Y.; Hou, Y.; Li, Q.; Guo, Y.; Feng, B.; Cui, L.; Wang, X.; et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J. Cell Mol. Med. 2017, 21, 1803–1814. [Google Scholar] [CrossRef]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.-C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Chen, G.; Lv, F.; Liu, Y.; Tian, H.; Tao, R.; Jiang, R.; Zhang, W.; Zhuo, C. LncRNA TINCR attenuates cardiac hypertrophy by epigenetically silencing CaMKII. Oncotarg 2017, 8, 47565–47573. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, W.; Guo, Y.; Chen, W.; Zheng, P.; Zeng, J.; Tong, W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE 2017, 12, e0185406. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Aguilo, F.; Zhou, M.-M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369. [Google Scholar] [CrossRef]
- He, C.; Yang, W.; Yang, J.; Ding, J.; Li, S.; Wu, H.; Zhou, F.; Jiang, Y.; Teng, L.; Yang, J. Long Noncoding RNA MEG3 Negatively Regulates Proliferation and Angiogenesis in Vascular Endothelial Cells. DNA Cell Biol. 2017, 36, 475–481. [Google Scholar] [CrossRef]
- Bell, R.D.; Long, X.; Lin, M.; Bergmann, J.H.; Nanda, V.; Cowan, S.L.; Zhou, Q.; Han, Y.; Spector, D.L.; Zheng, D.; et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arter. Thromb. Vasc. Biol. 2014, 34, 1249–1259. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Hou, J.; Long, H.; Zhou, C.; Zheng, S.; Wu, H.; Guo, T.; Wu, Q.; Zhong, T.; Wang, T. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res. 2017, 8, 4. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xu, Y.; Wang, Z.; Wu, Y.; Chen, J.; Wang, G.; Lu, C.; Jia, W.; Xi, J.; Zhu, S.; et al. A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Cell Stem Cell 2018, 22, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Li, Y.; Zhou, L.; Cho, J.; Patel, B.; Terada, N.; Li, Y.; Bungert, J.; Qiu, Y.; Huang, S. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Rep. 2016, 14, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Müller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657. [Google Scholar] [CrossRef] [PubMed]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Lin, B.; Sheng, Y.; Yang, L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev. Cell 2017, 42, 333–348. [Google Scholar] [CrossRef]
- Kang, X.; Zhao, Y.; Van Arsdell, G.; Nelson, S.F.; Touma, M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA 2020, 26, 481–491. [Google Scholar] [CrossRef]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef]
- Carè, A.; Catalucci, D.; Felicetti, F.; Bonci, D.; Addario, A.; Gallo, P.; Bang, M.-L.; Segnalini, P.; Gu, Y.; Dalton, N.D.; et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007, 13, 613–618. [Google Scholar] [CrossRef]
- Zangrando, J.; Zhang, L.; Vausort, M.; Maskali, F.; Marie, P.-Y.; Wagner, D.R.; Devaux, Y. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genom. 2014, 15, 460. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, C.; Meng, M.; Tang, H. Long Noncoding RNA MHRT Protects Cardiomyocytes against H2O2-Induced Apoptosis. Biomol 2016, 24, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, R.; Vasan, R.S. Biomarkers in cardiovascular disease: Statistical assessment and section on key novel heart failure biomarkers. Trends CardioVasc. Med. 2017, 27, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Faida, O.; Said, A.; Samir, P.; Oteh, M.; Latiff, A.; Fadilah, A. NT-proBNP levels, as predictor of left ventricular systolic and diastolic dysfunction in patients with chronic heart failure. Int. J. Collab Res. Intern. Med. Public Health 2012, 4, 910–923. [Google Scholar]
- Paul, T.K.; Mukherjee, D. Silent myocardial infarction and risk of heart failure. Ann. Transl. Med. 2018, 6, S35. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Wang, T.; Zeng, L.; Zhu, S.; Cao, W.; Wu, W.; Wu, H.; Zou, T. Diagnostic potential of circulating LncRNAs in human cardiovascular disease: A meta-analysis. Biosci. Rep. 2018, 38, BSR20181610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Wang, Y.; Ding, H.; Xue, S.; Yu, H.; Hu, L.; Qi, H.; Wang, Y.; Zhu, W.; et al. KCNQ1OT1, HIF1A-AS2 and APOA1-AS are promising novel biomarkers for diagnosis of coronary artery disease. Clin. Exp. Pharm. Physiol. 2019, 46, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Li, K.; Liu, S.; Huang, Y.; Zhao, X.; Huang, L.; Wang, Z.; et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Sci. Rep. 2016, 6, 22384. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef]
- Wang, X.-M.; Li, X.-M.; Song, N.; Zhai, H.; Gao, X.-M.; Yang, Y.-N. Long non-coding RNAs H19, MALAT1 and MIAT as potential novel biomarkers for diagnosis of acute myocardial infarction. Biomed. Pharm. 2019, 118, 109208. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, W.; Long, Q.-Q.; Zhang, J.; Li, Y.-F.; Liu, D.-C.; Yan, J.-J.; Yang, Z.-J.; Wang, L.-S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017, 7, 7491. [Google Scholar] [CrossRef]
- Boeckel, J.-N.; Perret, M.F.; Glaser, S.F.; Seeger, T.; Heumüller, A.W.; Chen, W.; John, D.; Kokot, K.E.; Katus, H.A.; Haas, J.; et al. Identification and regulation of the long non-coding RNA Heat2 in heart failure. J. Mol. Cell Cardiol. 2019, 126, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long Noncoding RNAs in Patients With Acute Myocardial Infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; Groote, P.d.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients With Heart Failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Y.-F.; Yang, X.-C.; Xu, L.; Li, W.-M.; Xia, K.; Zhang, D.-P.; Wu, R.-N.; Gan, T. Circulating Long Noncoding RNA LIPCAR Acts as a Novel Biomarker in Patients with ST-Segment Elevation Myocardial Infarction. Med. Sci. Monit. 2018, 24, 5064–5070. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, N.; Luo, P.; Jing, W.; Wen, X.; Liang, C.; Tu, J. Peripheral Blood Leukocyte Expression of lncRNA MIAT and Its Diagnostic and Prognostic Value in Ischemic Stroke. J. Stroke Cereb. Dis. 2018, 27, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016, 14, 183. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Y.-J.; Zhang, S.-L. Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J. Geriatr. Cardiol. 2019, 16, 818–821. [Google Scholar] [PubMed]
- Yan, Y.; Zhang, B.; Liu, N.; Qi, C.; Xiao, Y.; Tian, X.; Li, T.; Liu, B. Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction. Biomed. Res. Int. 2016, 2016, 8079372. [Google Scholar] [CrossRef]
- Kontaraki, J.E.; Marketou, M.E.; Kochiadakis, G.E.; Maragkoudakis, S.; Konstantinou, J.; Vardas, P.E.; Parthenakis, F.I. The long non-coding RNAs MHRT, FENDRR and CARMEN, their expression levels in peripheral blood mononuclear cells in patients with essential hypertension and their relation to heart hypertrophy. Clin. Exp. Pharm. Physiol. 2018, 45, 1213–1217. [Google Scholar] [CrossRef]
- Zhang, M.; Gu, H.; Xu, W.; Zhou, X. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int. J. Cardiol. 2016, 203, 214–216. [Google Scholar] [CrossRef]
- De Gonzalo-Calvo, D.; Kenneweg, F.; Bang, C.; Toro, R.; van der Meer, R.W.; Rijzewijk, L.J.; Smit, J.W.; Lamb, H.J.; Llorente-Cortes, V.; Thum, T. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 2016, 6, 3735. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).