In-Series U-Net Network to 3D Tumor Image Reconstruction for Liver Hepatocellular Carcinoma Recognition
Abstract
:1. Introduction
2. Proposed Method
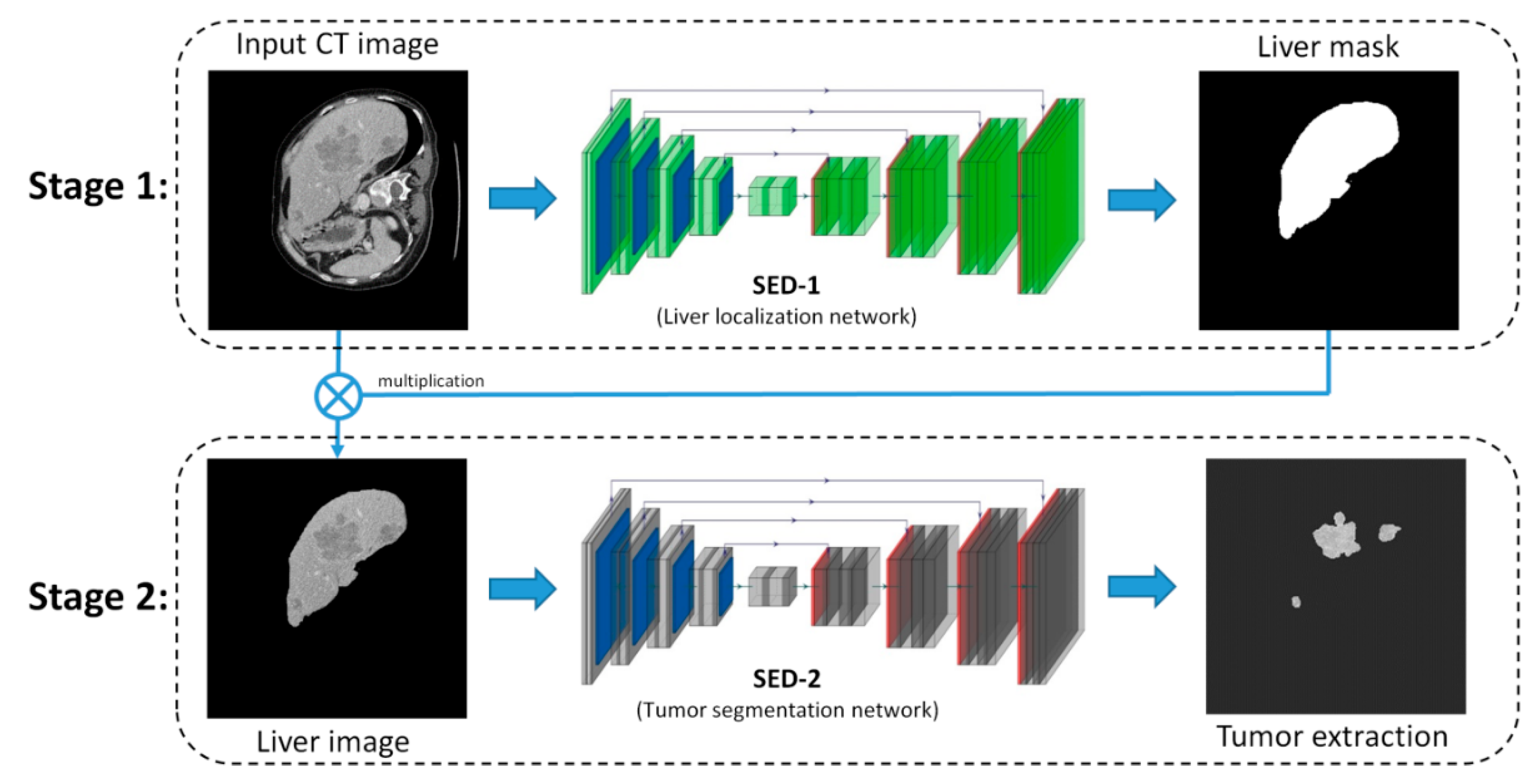
2.1. The Overview of SED
2.2. SED-1: Liver Localization Network
2.3. SED-2: Tumor Extraction Network
2.4. Loss Function
3. Liver CT Dataset
4. Results and Discussion
4.1. Training Method, Environment, and Parameter Setting
4.2. Evaluation Metrics
4.3. Tumor Segmentatiuon Results
- AUC = 0.5 (no discrimination);
- 0.7 ≤ AUC ≤ 0.8 (acceptable discrimination);
- 0.8 ≤ AUC ≤ 0.9 (excellent discrimination);
- 0.9 ≤ AUC ≤ 1.0 (outstanding discrimination).
4.4. 3D Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echegaray, S.; Gevaert, O.; Shah, R.; Kamaya, A.; Louie, J.; Kothary, N.; Napel, S. Core samples for radiomics features that are insensitive to tumor segmentation: Method and pilot study using CT images of hepatocellular carcinoma. J. Med. Imaging 2015, 2, 041011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Xie, L.; Fishman, E.K.; Yuille, A.L. Deep supervision for pancreatic cyst segmentation in abdominal CT scans. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; pp. 222–230. [Google Scholar]
- Shan, H.; Padole, A.; Homayounieh, F.; Kruger, U.; Khera, R.D.; Nitiwarangkul, C.; Kalra, M.K.; Wang, G. Can deep learning outperform modern commercial CT image reconstruction methods? Nat. Mach. Intell. 2018, 1, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Stefano, A.; Comelli, A.; Bravatà, V.; Barone, S.; Daskalovski, I.; Savoca, G.; Sabini, M.G.; Ippolito, M.; Russo, G. A preliminary PET radiomics study of brain metastases using a fully automatic segmentation method. BMC Bioinform. 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Comelli, A.; Dahiya, N.; Stefano, A.; Benfante, V.; Gentile, G.; Agnese, V.; Raffa, G.M.; Pilato, M.; Yezzi, A.; Petrucci, G.; et al. Deep learning approach for the segmentation of aneurysmal ascending aorta. Biomed. Eng. Lett. 2020, 1–10. [Google Scholar] [CrossRef]
- Klinder, T.; Ostermann, J.; Ehm, M.; Franz, A.; Kneser, R.; Lorenz, C. Automated model-based vertebra detection, identification, and segmentation in CT images. Med. Image Anal. 2009, 13, 471–482. [Google Scholar] [CrossRef]
- Varma, V.; Mehta, N.; Kumaran, V.; Nundy, S. Indications and contraindications for liver transplantation. Int. J. Hepatol. 2011, 2011, 121862. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.M.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Marziliano, P.; Thng, C.H. Liver tumor volume estimation by semi-automatic segmentation method. In Proceedings of the IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 31 August–3 September 2005. [Google Scholar]
- Lee, N.; Laine, A.F.; Klein, A. Towards a deep learning approach to brain parcellation. In Proceedings of the IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015. [Google Scholar]
- Li, W.; Jia, F.; Hu, Q. Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J. Comput. Commun. 2015, 3, 146–151. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Xie, W.; Zhang, Y.; Lu, Y.J.I.A. RIC-Unet: An Improved Neural Network Based on Unet for Nuclei Segmentation in Histology Images. IEEE Access 2019, 7, 21420–21428. [Google Scholar] [CrossRef]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Shen, D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- SAITO, K.; Huimin, L.; Hyoungseop, K.; Shoji, K.; Tanabe, M. ROI-based Fully Automated Liver Registration in Multi-phase CT Images. In Proceedings of the 18th International Conference on Control, Automation and Systems (ICCAS), Daegwallyeong, Korea, 17–20 October 2018. [Google Scholar]
- Hu, J.; Wang, H.; Gao, S.; Bao, M.; Liu, T.; Wang, Y.; Zhang, J. S-UNet: A Bridge-Style U-Net Framework with a Saliency Mechanism for Retinal Vessel Segmentation. IEEE Access 2019, 7, 174167–174177. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar]
- Zheng, Y.T. The Segmentation of Liver and Lesion Using Fully Convolution Neural Networks. Master’s Thesis, National University of Kaohsiung, Kaohsiung, Taiwan, 2017. [Google Scholar]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Gruber, N.; Antholzer, S.; Jaschke, W.; Kremser, C.; Haltmeier, M. A Joint Deep Learning Approach for Automated Liver and Tumor Segmentation. In Proceedings of the 13th International conference on Sampling Theory and Applications (SampTA), Bordeaux, France, 8–12 July 2019. [Google Scholar]
- Chlebus, G.; Meine, H.; Moltz, J.H.; Schenk, A. Neural Network-Based Automatic Liver Tumor Segmentation with Random Forest-Based Candidate Filtering. arXiv 2017, arXiv:1706.00842. [Google Scholar]
- Han, X. Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv 2017, arXiv:abs/1704.07239. [Google Scholar]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the Diagnosis of Diabetic and Hypertensive Retinopathy Using Artificial Intelligence-Based Semantic Segmentation. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [Green Version]
- Ünver, H.M.; Ayan, E. Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and Grab Cut Algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Xue, Z.; Rodney Long, L.; Antani, S. Cross-Dataset Evaluation of Deep Learning Networks for Uterine Cervix Segmentation. Diagnostics 2020, 10, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjdira, B.; Ouni, K.; Al Rahhal, M.M.; Albakr, A.; Al-Habib, A.; Mahrous, E. Spinal Cord Segmentation in Ultrasound Medical Imagery. Appl. Sci. 2020, 10, 1370. [Google Scholar] [CrossRef] [Green Version]
- Kolařík, M.; Burget, R.; Uher, V.; Říha, K.; Dutta, M.K. Optimized High Resolution 3D Dense-U-Net Network for Brain and Spine Segmentation. Appl. Sci. 2019, 9, 404. [Google Scholar] [CrossRef] [Green Version]
- El Adoui, M.; Mahmoudi, S.A.; Larhmam, M.A.; Benjelloun, M. MRI Breast Tumor Segmentation Using Different Encoder and Decoder CNN Architectures. Computers 2019, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Gadosey, P.K.; Li, Y.; Agyekum, E.A.; Zhang, T.; Liu, Z.; Yamak, T.; Essaf, F. SD-UNet: Stripping Down U-Net for Segmentation of Biomedical Images on Platforms with Low Computational Budgets. Diagnostics 2020, 10, 110. [Google Scholar] [CrossRef] [Green Version]
- Iesmantas, T.; Paulauskaite-Taraseviciene, A.; Sutiene, K. Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning. Appl. Sci. 2020, 10, 615. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Lian, S.; Zhimimg, L.; Li, S. Weighted Res-UNet for High-quality Retina Vessel Segmentation. In Proceedings of the 9th International Conference on Information Technology in Medicine and Education, Hangzhou, China, 19–21 October 2018; Volume 10, pp. 328–331. [Google Scholar]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Huang, G.; Liu, Z.; Vazquez, D.; van der Maaten, L. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef] [Green Version]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer Science+ Business Media: New York, NY, USA, 2006. [Google Scholar]
- Collins, M.; Schapire, R.E.; Singer, Y. Logistic regression, AdaBoost and Bregman distances. Mach. Learn. 2002, 48, 253–285. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F.; Pal, S. Keras; Packt: Birmingham, UK, 2015. [Google Scholar]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, E.C., Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A guide for authors and reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [Green Version]
- Furey, T.S.; Cristianini, N.; Duffy, N.; Bednarski, D.W.; Schummer, M.; Haussler, D.J.B. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 2000, 16, 906–914. [Google Scholar] [CrossRef]
- Comelli, A.; Coronnello, C.; Dahiya, N.; Benfante, V.; Palmucci, S.; Basile, A.; Vancheri, C.; Russo, G.; Yezzi, A.; Stefano, A. Lung Segmentation on High-Resolution Computerized Tomography Images Using Deep Learning: A Preliminary Step for Radiomics Studies. J. Imaging 2020, 6, 125. [Google Scholar] [CrossRef]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. Enet: A deep neural network architecture for real-time semantic segmentation. arXiv 2016, arXiv:1606.02147. [Google Scholar]
- Santoro, A.; Bartunov, S.; Botvinick, M.; Wierstra, D.; Lillicrap, T. One-shot learning with memory-augmented neural networks. arXiv 2016, arXiv:1605.06065. [Google Scholar]
- Lits-Challenge. Available online: https://competitions.codalab.org/competitions/17094 (accessed on 4 August 2017).












| Encoder | Output Size | Decoder | Connecting Operation | Output Size |
|---|---|---|---|---|
| Input | 256^2 × 1 | UP 1 | 32^2 × 256 | |
| Conv. block 1 | 256^2 × 32 | Copy 1 | [Conv. block 4] | 32^2 × 256 |
| Pooling | 128^2 × 32 | Conv. block 6 | [UP1, Copy1] | 32^2 × 128 |
| Conv. block 2 | 128^2 × 64 | UP 2 | 64^2 × 128 | |
| Pooling | 64^2 × 64 | Copy 2 | [Conv. block 3] | 64^2 × 128 |
| Conv. block 3 | 64^2 × 128 | Conv. block 7 | [UP2, Copy 2] | 64^2 × 64 |
| Pooling | 32^2 × 128 | UP 3 | 128^2 × 64 | |
| Conv. block 4 | 32^2 × 256 | Copy 3 | [Conv. block 2] | 128^2 × 64 |
| Pooling | 16^2 × 256 | Conv. block 8 | [UP3, Copy 3] | 128^2 × 32 |
| Conv. block 5 | 16^2 × 512 | UP 4 | 256^2 × 32 | |
| Copy 4 | [Conv. block 1] | 256^2 × 32 | ||
| Conv. block 9 | [UP4, Copy 4] | 256^2 × 16 | ||
| Conv. | 256^2 × 1 |
| Encoder | Output Size | Decoder | Connecting Operation | Output Size |
|---|---|---|---|---|
| Input | 256^2 × 1 | TU 1 | 16^2 × 240 | |
| Conv | 256^2 × 48 | Copy 1 | [DB 5] | 16^2 × 656 |
| DB 1 (4 layers) | 256^2 × 112 | DB 7 (12 layers) | [TU 1, Copy 1] | 16^2 × 192 |
| TD 1 | 128^2 × 112 | TU 2 | 32^2 × 192 | |
| DB 2 (5 layers) | 128^2 × 192 | Copy 2 | [DB 4] | 32^2 × 464 |
| TD 2 | 64^2 × 192 | DB 8 (10 layers) | [TU 2, Copy 2] | 32^2 × 160 |
| DB 3 (7 layers) | 64^2 × 304 | TU 3 | 64^2 × 160 | |
| TD 3 | 32^2 × 304 | Copy 3 | [DB 3] | 64^2 × 304 |
| DB 4 (10 layers) | 32^2 × 464 | DB 9 (7 layers) | [TU 3, Copy 3] | 64^2 × 112 |
| TD 4 | 16^2 × 464 | TU 4 | 128^2 × 112 | |
| DB 5 (12 layers) | 16^2 × 656 | Copy 4 | [DB 2] | 128^2 × 192 |
| TD 5 | 8^2 × 656 | DB 10 (5 layers) | [TU 4, Copy 4] | 128^2 × 80 |
| DB 6 (15 layers) | 8^2 × 880 | TU 5 | 256^2 × 80 | |
| Copy 5 | [DB 1] | 256^2 × 112 | ||
| DB 11 (4 layers) | [TU 5, Copy 5] | 256^2 × 1 |
| Methods | ACC | IoU | DSC | AUC |
|---|---|---|---|---|
| U-Net [19] | 0.92 | 0.53 | 0.65 | 0.73 |
| ResNet [29] | 0.98 | 0.62 | 0.67 | 0.77 |
| C-UNet [27] | 0.99 | 0.67 | 0.67 | 0.87 |
| Our SED | 0.992 | 0.87 | 0.75 | 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.-F.; Ou, H.-Y.; Liu, K.-H.; Li, Z.-Y.; Liao, C.-C.; Wang, S.-Y.; Huang, W.; Cheng, Y.-F.; Pan, C.-T. In-Series U-Net Network to 3D Tumor Image Reconstruction for Liver Hepatocellular Carcinoma Recognition. Diagnostics 2021, 11, 11. https://doi.org/10.3390/diagnostics11010011
Chen W-F, Ou H-Y, Liu K-H, Li Z-Y, Liao C-C, Wang S-Y, Huang W, Cheng Y-F, Pan C-T. In-Series U-Net Network to 3D Tumor Image Reconstruction for Liver Hepatocellular Carcinoma Recognition. Diagnostics. 2021; 11(1):11. https://doi.org/10.3390/diagnostics11010011
Chicago/Turabian StyleChen, Wen-Fan, Hsin-You Ou, Keng-Hao Liu, Zhi-Yun Li, Chien-Chang Liao, Shao-Yu Wang, Wen Huang, Yu-Fan Cheng, and Cheng-Tang Pan. 2021. "In-Series U-Net Network to 3D Tumor Image Reconstruction for Liver Hepatocellular Carcinoma Recognition" Diagnostics 11, no. 1: 11. https://doi.org/10.3390/diagnostics11010011
APA StyleChen, W.-F., Ou, H.-Y., Liu, K.-H., Li, Z.-Y., Liao, C.-C., Wang, S.-Y., Huang, W., Cheng, Y.-F., & Pan, C.-T. (2021). In-Series U-Net Network to 3D Tumor Image Reconstruction for Liver Hepatocellular Carcinoma Recognition. Diagnostics, 11(1), 11. https://doi.org/10.3390/diagnostics11010011






