Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters
Abstract
1. Introduction
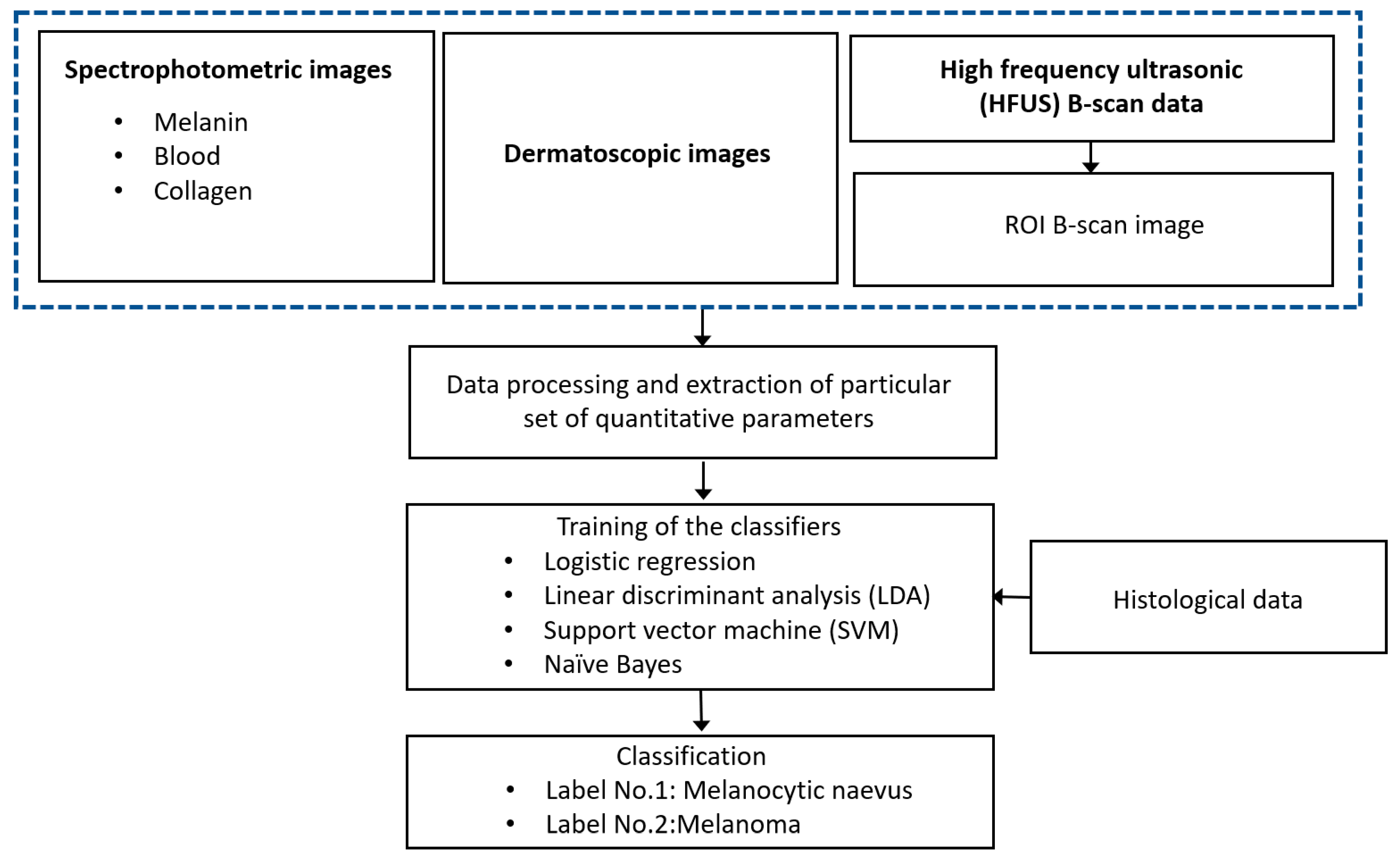
2. Description of the Proposed Technique
3. Experimental Analysis and Clinical Measurements
4. Data Processing
5. Classification Algorithm
6. Results and Discussion
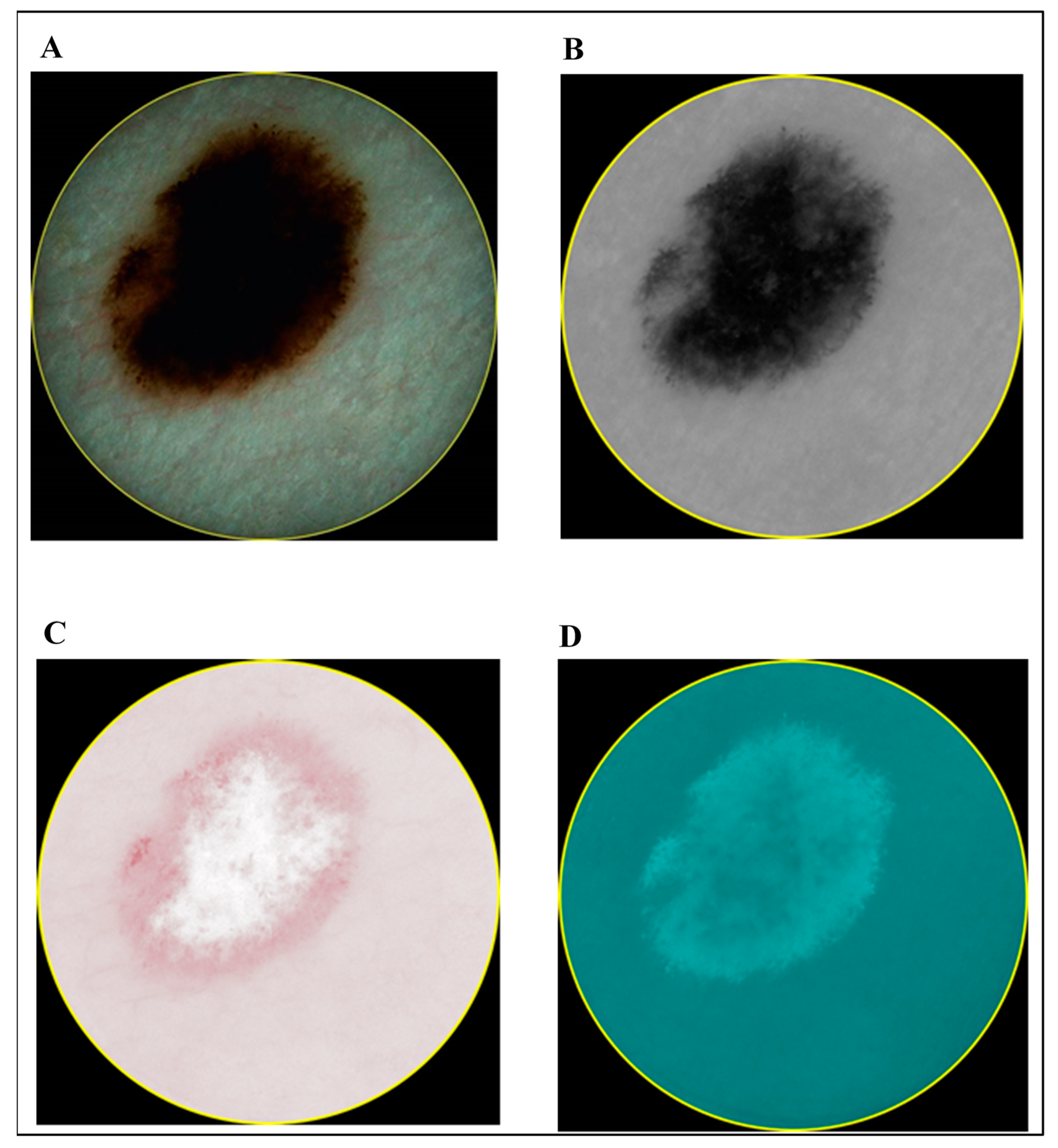
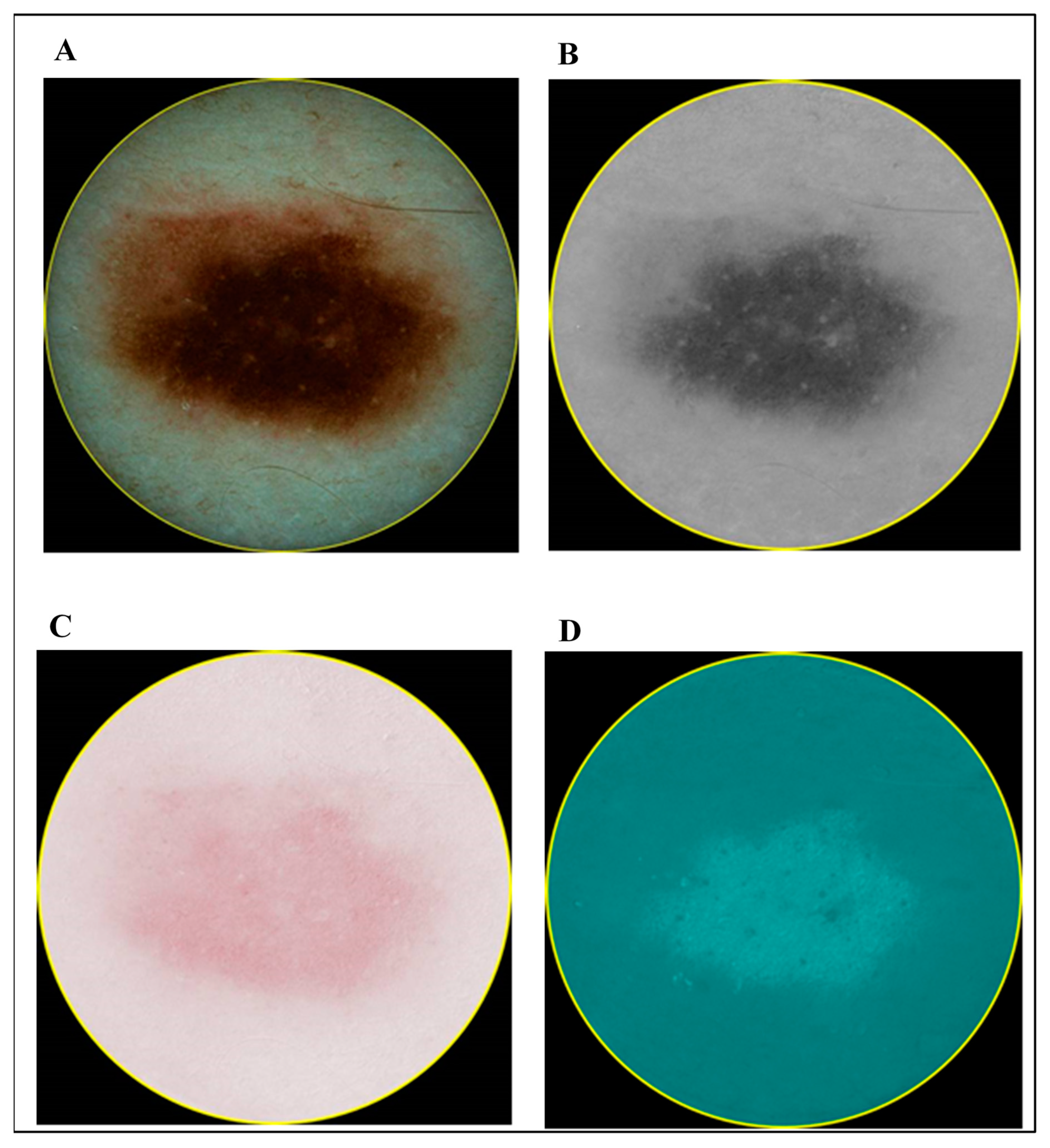
- Case 1: Combining Quantitative Parameters from Dermatoscopic and Spectrophotometric Images
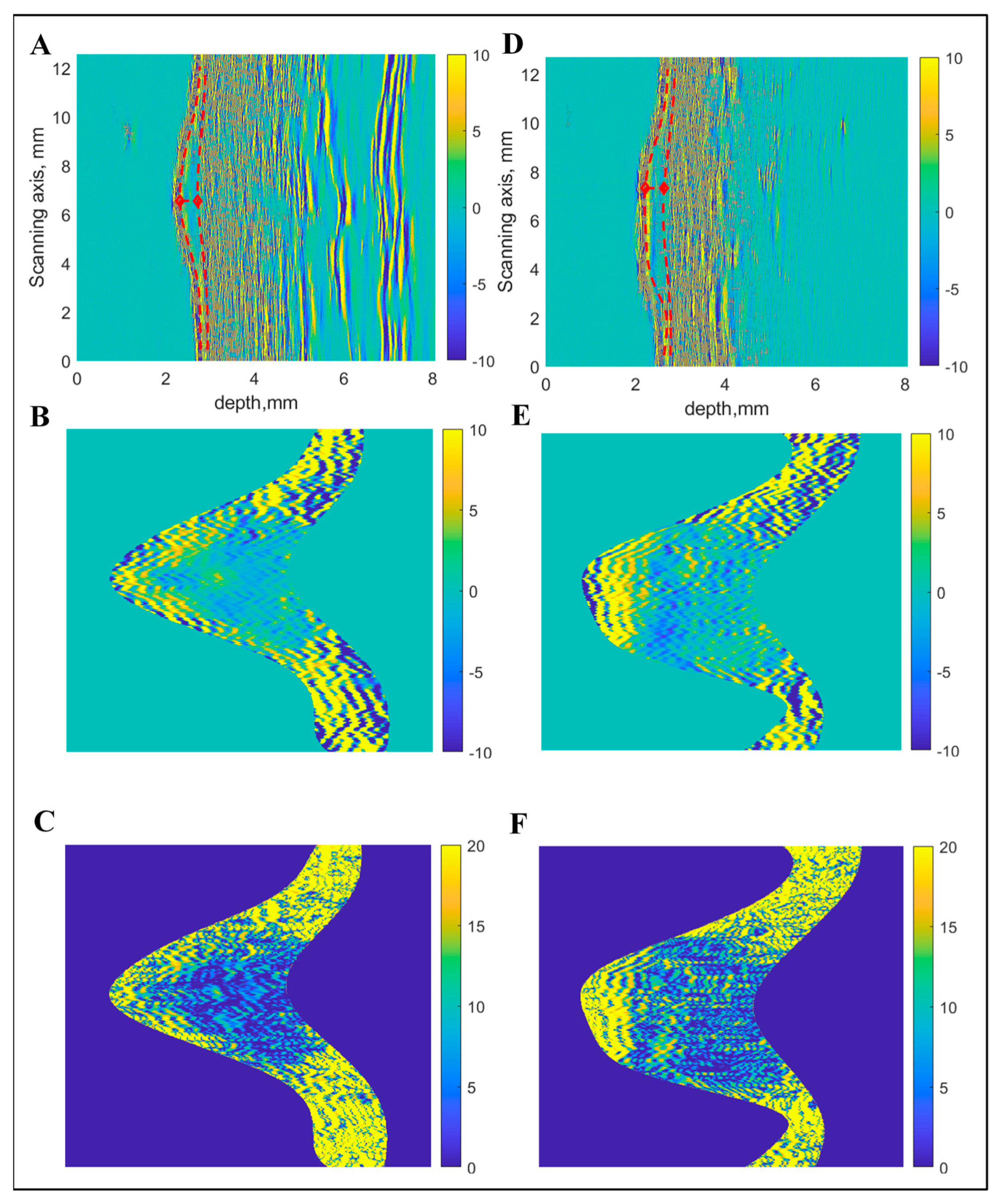
- Case 2: Combining Quantitative Parameters from Dermatoscopic and Ultrasonic B-scan Images
- Case 3: Combination of Spectrophotometry and HFUS Imaging Techniques
- Case 4: Combination of All Three (Optical Dermatoscopy, Spectrophotometry and HFUS) Imaging Techniques
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Force, U.P.S.T.; Bibbins-Domingo, K.; Grossman, D.C.; Curry, S.J.; Davidson, K.W.; Ebell, M.; Epling, J.W.; García, F.A.R.; Gillman, M.W.; Kemper, A.R.; et al. Screening for Skin Cancer. JAMA 2016, 316, 429–435. [Google Scholar] [CrossRef]
- Gardner, L.J.; Strunck, J.L.; Wu, Y.P.; Grossman, D. Current controversies in early-stage melanoma: Questions on incidence, screening, and histologic regression. J. Am. Acad. Dermatol. 2019, 80, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Watts, C.G.; Dieng, M.; Morton, R.L.; Mann, G.J.; Menzies, S.W.; Cust, A.E. Clinical practice guidelines for identification, screening and follow-up of individuals at high risk of primary cutaneous melanoma: A systematic review. Br. J. Dermatol. 2014, 172, 33–47. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. For members of the American Joint Committee on Cancer Melanoma Expert Panel and the International Melanoma Database and Discovery Platform Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar]
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Swetter, S.M.; Tsao, H.; Bichakjian, C.K.; Curiel-Lewandrowski, C.; Elder, D.E.; Gershenwald, J.E.; Guild, V.; Grant-Kels, J.M.; Halpern, A.C.; Johnson, T.M.; et al. Guidelines of care for the management of primary cutaneous melanoma. J. Am. Acad. Dermatol. 2019, 80, 208–250. [Google Scholar] [CrossRef]
- Vestergaard, M.E.; Macaskill, P.; Holt, P.E.; Menzies, S.W. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta-analysis of studies performed in a clinical setting. Br. J. Dermatol. 2008, 159, 669–676. [Google Scholar] [CrossRef]
- Salerni, G.; Terán, T.; Puig, S.; Malvehy, J.; Zalaudek, I.; Argenziano, G.; Kittler, H. Meta-analysis of digital dermoscopy follow-up of melanocytic skin lesions: A study on behalf of the International Dermoscopy Society. J. Eur. Acad. Dermatol. Venereol. 2012, 27, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, A.; Silvers, D. Dermatopathology standards. J. Cutan. Pathol. 1996, 23, 194–195. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.R.; Gonin, R.; Hanna, M.P. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum. Pathol. 1996, 27, 528–531. [Google Scholar] [CrossRef]
- Elmore, J.G.; Barnhill, R.L.; Elder, D.E.; Longton, G.M.; Pepe, M.S.; Reisch, L.M.; Carney, P.A.; Titus, L.J.; Nelson, H.D.; Onega, T.; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357, j2813. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Berking, C.; Klode, J.; Schadendorf, D.; Jansen, P.; Franklin, C.; Holland-Letz, T.; Krahl, D.; et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur. J. Cancer 2019, 115, 79–83. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems., A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Fröhling, S.; et al. Collaborators A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur. J. Cancer 2019, 111, 148–154. [Google Scholar] [CrossRef]
- Dick, V.; Sinz, C.; Mittlböck, M.; Kittler, H.; Tschandl, P. Accuracy of Computer-Aided Diagnosis of Melanoma: A Meta-analysis. JAMA Dermatol. 2019, 155, 1291–1299. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Cui, X.; Wei, R.; Gong, L.; Qi, R.; Zhao, Z.; Chen, H.; Song, K.; Abdulrahman, A.A.A.; Wang, Y.; Chen, J.Z.S.; et al. Assessing the effectiveness of artificial intelligence methods for melanoma: A retrospective review. J. Am. Acad. Dermatol. 2019, 81, 1176–1180. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Collaborators Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ferrante di Ruffano, L.; Takwoingi, Y.; Dinnes, J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Matin, R.N.; Godfrey, K.; O’Sullivan, C.; Gulati, A.; et al. Cochrane Skin Cancer Diagnostic Test Accuracy Group Computer-assisted diagnosis techniques (dermoscopy and spectroscopy-based) for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013186. [Google Scholar] [PubMed]
- Schneider, S.L.; Kohli, I.; Hamzavi, I.H.; Council, M.L.; Rossi, A.M.; Ozog, D.M. Emerging imaging technologies in dermatology: Part I: Basic principles. J. Am. Acad. Dermatol. 2019, 80, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.L.; Kohli, I.; Hamzavi, I.H.; Council, M.L.; Rossi, A.M.; Ozog, D.M. Emerging imaging technologies in dermatology: Part II: Applications and limitations. J. Am. Acad. Dermatol. 2019, 80, 1121–1131. [Google Scholar] [CrossRef]
- Pellacani, G.; Witkowski, A.; Cesinaro, A.M.; Losi, A.; Colombo, G.L.; Campagna, A.; Longo, C.; Piana, S.; de Carvalho, N.; Giusti, F.; et al. Cost-benefit of reflectance confocal microscopy in the diagnostic performance of melanoma. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 413–419. [Google Scholar] [CrossRef]
- Mataca, E.; Migaldi, M.; Cesinaro, A.M. Impact of Dermoscopy and Reflectance Confocal Microscopy on the Histopathologic Diagnosis of Lentigo Maligna/Lentigo Maligna Melanoma. Am. J. Dermatopathol. 2018, 40, 884–889. [Google Scholar] [CrossRef]
- Sakalauskienė, K.; Valiukevičienė, S.; Raišutis, R.; Linkevičiūtė, G. The significance of spectrophotometric image analysis for diagnosis of the melanocytic skin tumours in association with their thickness. Skin Res. Technol. 2018, 24, 692–698. [Google Scholar] [CrossRef]
- Walter, F.M.; Morris, H.C.; Humphrys, E.; Hall, P.N.; Prevost, A.T.; Burrows, N.; Bradshaw, L.; Wilson, E.C.F.; Norris, P.; Walls, J.; et al. Effect of adding a diagnostic aid to best practice to manage suspicious pigmented lesions in primary care: Randomised controlled trial. BMJ 2012, 345, e4110. [Google Scholar] [CrossRef]
- Kleinerman, R.; Whang, T.B.; Bard, R.L.; Marmur, E.S. Ultrasound in dermatology: Principles and applications. J. Am. Acad. Dermatol. 2012, 67, 478–487. [Google Scholar] [CrossRef]
- Jasaitiene, D.; Valiukeviciene, S.; Linkeviciute, G.; Raisutis, R.; Jasiuniene, E.; Kazys, R. Principles of high-frequency ultrasonography for investigation of skin pathology. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Chaput, L.; Laurent, E.; Pare, A.; Sallot, A.; Mourtada, Y.; Ossant, F.; Vaillant, L.; Patat, F.; Machet, L. One-Step surgical removal of cutaneous melanoma with surgical margins based on preoperative ultrasound measurement of the thickness of the melanoma. Eur. J. Dermatol. 2018, 28, 202–208. [Google Scholar] [CrossRef]
- Dinnes, J.; Bamber, J.; Chuchu, N.; Bayliss, S.E.; Takwoingi, Y.; Davenport, C.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; Deeks, J.J.; et al. Cochrane Skin Cancer Diagnostic Test Accuracy Group High-frequency ultrasound for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013188. [Google Scholar] [PubMed]
- Andrėkutė, K.; Linkevičiūtė, G.; Raišutis, R.; Valiukevičienė, S.; Makštienė, J. Automatic Differential Diagnosis of Melanocytic Skin Tumors Using Ultrasound Data. Ultrasound Med. Biol. 2016, 42, 2834–2843. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.T.; Defraene, G.; de Ruysscher, D.; Lambin, P.; van Elmpt, W. Quantitative radiomics studies for tissue characterization: A review of technology and methodological procedures. Br. J. Radiol. 2017, 90, 20160665. [Google Scholar] [CrossRef]
- Yao, Z.; Dong, Y.; Wu, G.; Zhang, Q.; Yang, D.; Yu, J.H.; Wang, W.P. Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer 2018, 18, 1089. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, Y.; Qiao, M.; Wang, Y.; Yu, J.; Li, J.; Chang, C. Radiomics Analysis on Ultrasound for Prediction of Biologic Behavior in Breast Invasive Ductal Carcinoma. Clin. Breast Cancer 2018, 18, e335–e344. [Google Scholar] [CrossRef]
- Hu, H.; Wang, Z.; Huang, X.; Chen, S.; Zheng, X.; Ruan, S.; Xie, X.; Lu, M.; Yu, J.; Tian, J.; et al. Ultrasound-based radiomics score: A potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur. Radiol. 2019, 29, 2890–2901. [Google Scholar] [CrossRef]
- Drulyte, I.; Ruzgas, T.; Raisutis, R.; Valiukeviciene, S.; Linkeviciute, G. Application of automatic statistical post-processing method for analysis of ultrasonic and digital dermatoscopy images. Libyan J. Med. 2018, 13, 1479600. [Google Scholar] [CrossRef]
- Andrekute, K.; Valiukeviciene, S.; Raisutis, R.; Linkeviciute, G.; Makstiene, J.; Kliunkiene, R. Automated Estimation of Melanocytic Skin Tumor Thickness by Ultrasonic Radiofrequency Data. J. Ultrasound Med. 2016, 35, 857–865. [Google Scholar] [CrossRef]
- Rey-Barroso, L.; Burgos-Fernández, F.J.; Delpueyo, X.; Ares, M.; Royo, S.; Malvehy, J.; Puig, S.; Vilaseca, M. Visible and Extended Near-Infrared Multispectral Imaging for Skin Cancer Diagnosis. Sensors 2018, 18, 1441. [Google Scholar] [CrossRef] [PubMed]
- Echegaray, S.; Bakr, S.; Rubin, D.L.; Napel, S. Quantitative Image Feature Engine (QIFE): An Open-Source, Modular Engine for 3D Quantitative Feature Extraction from Volumetric Medical Images. J. Digit. Imaging 2018, 31, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Kotsiantis, S.B.; Zaharakis, I.D.; Pintelas, P.E. Machine learning: A review of classification and combining techniques. Artif. Intell. Rev. 2006, 26, 159–190. [Google Scholar] [CrossRef]
- Shi, Z.; He, L.; Suzuki, K.; Nakamura, T.; Itoh, H. Survey on Neural Networks Used for Medical Image Processing. Int. J. Comput. Sci. 2009, 3, 86–100. [Google Scholar]
- Pisner, D.A.; Schnyer, D.M. Chapter 6-Support vector machine. In Machine Learning; Mechelli, A., Vieira, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 101–121. [Google Scholar]
- Hiam, A.; Eman, S.; Michael, O.J.; Mufeed, M. Enhancement of 3D modeling and classification of microcalcifications in breast computed tomography (BCT). In Proceedings of the Society of Photo-Optical Instrumentation Engineers (SPIE) Conference Series, San Diego, CA, USA, 21 March 2014; p. 903436. [Google Scholar]
- Whiteman, D.C.; Thompson, B.S.; Thrift, A.P.; Hughes, M.; Muranushi, C.; Neale, R.E.; Green, A.C.; Olsen, C.M.; Whiteman, D.C.; Green, A.C.; et al. A Model to Predict the Risk of Keratinocyte Carcinomas. J. Investig. Dermatol. 2016, 136, 1247–1254. [Google Scholar] [CrossRef]
- Mohanty, N.; John, A.L.; Manmatha, R.; Rath, T.M. Chapter 10-Shape-Based Image Classification and Retrieval. Handb. Stat. 2013, 31, 249–267. [Google Scholar] [CrossRef]
- Liu, C. Discriminant analysis and similarity measure. Pattern Recognit. 2014, 47, 359–367. [Google Scholar] [CrossRef]
- Jadhav, P.; Guru, S.K. Image classification using naive bayes model for deep head pose estimation. Int. J. Adv. Eng. Res. Dev. 2017, 4, 594–599. [Google Scholar]
- Jiang, L.; Zhang, H.; Cai, Z. A Novel Bayes Model: Hidden Naive Bayes. IEEE Trans. Knowl. Data Eng. 2009, 21, 1361–1371. [Google Scholar] [CrossRef]
- Bhalla, S.; Kaur, H.; Dhall, A.; Raghava, G.P.S. Prediction and Analysis of Skin Cancer Progression using Genomics Profiles of Patients. Sci. Rep. 2019, 9, 15790. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, M. Support Vector Machines: A Tutorial Overview and Critical Appraisal. In Proceedings of the IEE Colloquium on Applied Statistical Pattern Recognition 1999, Brimingham, UK, 20 April 1999; pp. 1–2. [Google Scholar] [CrossRef]
- Murugan, A.; Nair, S.A.; Kumar, K.P.S. Detection of Skin Cancer Using SVM, Random Forest and kNN Classifiers. J. Med. Syst. 2019, 43, 1–9. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Arasi, M.A.; Dahshan, E.; Horbaty, S.M.; Salem, A.M. Malignant Melanoma Detection Based on Machine Learning Techniques: A Survey. Egypt. Comput. Sci. J. 2016, 40, 1–10. [Google Scholar]
- Seeja, R.D.; Suresh, A. Deep Learning Based Skin Lesion Segmentation and Classification of Melanoma Using Support Vector Machine (SVM). Asian Pac. J. Cancer Prev. 2019, 20, 1555–1561. [Google Scholar] [CrossRef]
- Alquran, H.; Abu Qasmieh, I.; Alqudah, A.M.; Alhammouri, S.; Alawneh, E.; AbuGhazaleh, A.; Hasayen, F. The melanoma skin cancer detection and classification using support vector machine. In Proceedings of the 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT), Amman, Jordan, 11–13 October 2017; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2017; pp. 1–5. [Google Scholar]
- Patil, M.; Dongre, N. Melanoma Detection Using HSV with SVM Classifier and De-duplication Technique to Increase Efficiency. Commun. Comput. Inf. Sci. 2020, 109–120. [Google Scholar] [CrossRef]
- Jaworek-Korjakowska, J. Computer-Aided Diagnosis of Micro-Malignant Melanoma Lesions Applying Support Vector Machines. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Bakheet, S. An SVM Framework for Malignant Melanoma Detection Based on Optimized HOG Features. Computation 2017, 5, 4. [Google Scholar] [CrossRef]
| Type of Imaging Technology and Images | Numbers of Selected Quantitative Parameters (p < 0.05) to be Used for Classification | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| Optical dermatoscopy | x | x | x | x | |||||||||
| Spectrophotometry (melanin component) | x | x | x | ||||||||||
| Spectrophotometry (blood component) | x | x | x | x | x | x | x | ||||||
| Spectrophotometry (collagen component) | x | x | x | x | |||||||||
| Ultrasonic B-scan | x | x | x | x | x | x | x | ||||||
| Type of Classifier | Statistical Parameters | |||||
|---|---|---|---|---|---|---|
| Accuracy, % | Sensitivity, % | Specificity, % | Precision, % | Matthews Correlation Coefficient (MCC) | Area under the ROC Curve | |
| Logistic regression (LR) | 89.01 | 85.37 | 92.00 | 89.74 | 0.778 | 0.918 |
| Linear discriminant analysis (LDA) | 84.62 | 78.05 | 90.00 | 86.49 | 0.689 | 0.906 |
| Support vector machine (SVM) | 90.11 | 85.37 | 94.0 | 92.11 | 0.801 | 0.972 |
| Naive Bayes | 74.73 | 58.54 | 88.00 | 80.00 | 0.493 | 0.813 |
| Type of Classifier | Statistical Parameters | |||||
|---|---|---|---|---|---|---|
| Accuracy, % | Sensitivity, % | Specificity, % | Precision, % | Matthews Correlation Coefficient (MCC) | Area under the ROC Curve | |
| Logistic regression (LR) | 82.42 | 78.05 | 86.00 | 82.05 | 0.644 | 0.908 |
| Linear discriminant analysis (LDA) | 80.22 | 73.17 | 86.00 | 81.08 | 0.599 | 0.906 |
| Support vector machine (SVM) | 91.21 | 80.49 | 100 | 100 | 0.833 | 0.961 |
| Naive Bayes | 76.92 | 75.61 | 78.00 | 73.81 | 0.535 | 0.812 |
| Type of Classifier | Statistical Parameters | |||||
|---|---|---|---|---|---|---|
| Accuracy, % | Sensitivity, % | Specificity, % | Precision, % | Matthews Correlation Coefficient (MCC) | Area under the ROC Curve | |
| Logistic regression (LR) | 85.71 | 85.37 | 86.00 | 83.33 | 0.712 | 0.928 |
| Linear discriminant analysis (LDA) | 86.81 | 85.37 | 88.00 | 85.37 | 0.734 | 0.905 |
| Support vector machine (SVM) | 95.60 | 92.68 | 98.00 | 97.44 | 0.912 | 0.996 |
| Naive Bayes | 73.63 | 65.85 | 80.00 | 72.97 | 0.465 | 0.82 |
| Type of Classifier | Statistical Parameters | |||||
|---|---|---|---|---|---|---|
| Accuracy, % | Sensitivity, % | Specificity, % | Precision, % | Matthews Correlation Coefficient (MCC) | Area under the ROC Curve | |
| Logistic regression (LR) | 92.31 | 87.80 | 96.00 | 94.74 | 0.846 | 0.956 |
| Linear discriminant analysis (LDA) | 90.11 | 85.37 | 94.00 | 92.11 | 0.801 | 0.939 |
| Support vector machine (SVM) | 98.9 | 97.56 | 100 | 100 | 0.978 | 0.999 |
| Naive Bayes | 75.82 | 65.85 | 84.00 | 77.14 | 0.51 | 0.829 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, K.A.; Raišutis, R.; Liutkus, J.; Valiukevičienė, S. Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters. Diagnostics 2020, 10, 632. https://doi.org/10.3390/diagnostics10090632
Tiwari KA, Raišutis R, Liutkus J, Valiukevičienė S. Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters. Diagnostics. 2020; 10(9):632. https://doi.org/10.3390/diagnostics10090632
Chicago/Turabian StyleTiwari, Kumar Anubhav, Renaldas Raišutis, Jokūbas Liutkus, and Skaidra Valiukevičienė. 2020. "Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters" Diagnostics 10, no. 9: 632. https://doi.org/10.3390/diagnostics10090632
APA StyleTiwari, K. A., Raišutis, R., Liutkus, J., & Valiukevičienė, S. (2020). Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters. Diagnostics, 10(9), 632. https://doi.org/10.3390/diagnostics10090632







