Machine Learning Model Comparison in the Screening of Cholangiocarcinoma Using Plasma Bile Acids Profiles
Abstract
1. Introduction
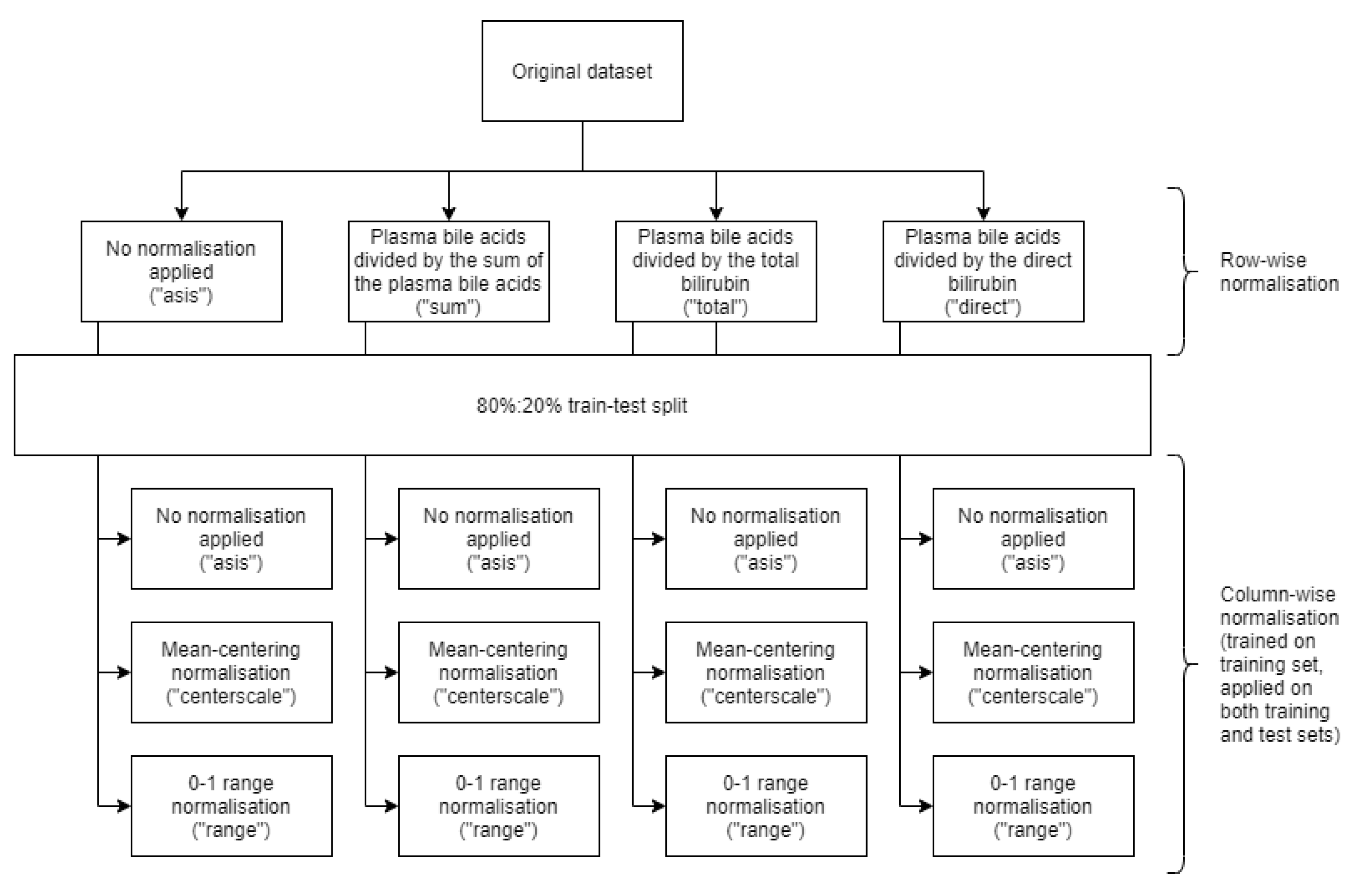
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BA (BAs) | Bile acid (Bile acids) |
| CCA | Cholangiocarcinoma |
| ML | Machine learning |
| AI | Artificial intelligence |
| UHPLC | Ultra (high) performance liquid chromatography |
| MS/MS | Tandem mass spectrometry |
| AUC | Area under curve |
| UMAP | Uniform manifold approximation and projection |
| ROC | Receiver operating characteristic |
References
- Lippi, G. Machine learning in laboratory diagnostics: Valuable resources or a big hoax? Diagnosis 2019. [Google Scholar] [CrossRef] [PubMed]
- Burke, H.B. Artificial neural networks for cancer research: Outcome prediction. Semin. Surg. Oncol. 1994, 10, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Salah, H.T.; Muhsen, I.N.; Salama, M.E.; Owaidah, T.; Hashmi, S.K. Machine learning applications in the diagnosis of leukemia: Current trends and future directions. Int. J. Lab. Hematol. 2019, 41, 717–725. [Google Scholar] [CrossRef] [PubMed]
- US FDA Digital Health Criteria. Available online: https://www.fda.gov/medical-devices/digital-health/digital-health-criteria (accessed on 13 July 2020).
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317. [Google Scholar] [CrossRef]
- Ghaffari, M.H.; Jahanbekam, A.; Sadri, H.; Schuh, K.; Dusel, G.; Prehn, C.; Adamski, J.; Koch, C.; Sauerwein, H. Metabolomics meets machine learning: Longitudinal metabolite profiling in serum of normal versus overconditioned cows and pathway analysis. J. Dairy Sci. 2019, 102, 11561–11585. [Google Scholar] [CrossRef]
- Gunčar, G.; Kukar, M.; Notar, M.; Brvar, M.; Černelč, P.; Notar, M.; Notar, M. An application of machine learning to haematological diagnosis. Sci. Rep. 2018, 8, 411. [Google Scholar] [CrossRef]
- Dreiseitl, S.; Ohno-Machado, L.; Kittler, H.; Vinterbo, S.; Billhardt, H.; Binder, M. A Comparison of Machine Learning Methods for the Diagnosis of Pigmented Skin Lesions. J. Biomed. Inform. 2001, 34, 28–36. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Danese, E.; Ruzzenente, A.; Montagnana, M.; Lievens, P.M.-J. Current and future roles of mucins in cholangiocarcinoma—recent evidences for a possible interplay with bile acids. Ann. Transl. Med. 2018, 6, 333. [Google Scholar] [CrossRef]
- Danese, E.; Salvagno, G.L.; Negrini, D.; Brocco, G.; Montagnana, M.; Lippi, G. Analytical evaluation of three enzymatic assays for measuring total bile acids in plasma using a fully automated clinical chemistry platform. PLoS ONE 2017, 12, e0179200. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Salvagno, G.L.; Tarperi, C.; Negrini, D.; Montagnana, M.; Festa, L.; Sanchis-Gomar, F.; Schena, F.; Lippi, G. Middle-distance running acutely influences the concentration and composition of serum bile acids: Potential implications for cancer risk? Oncotarget 2017, 8, 52775–52782. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Negrini, D.; Pucci, M.; De Nitto, S.; Ambrogi, D.; Donzelli, S.; Lievens, P.M.-J.; Salvagno, G.L.; Lippi, G. Bile Acids Quantification by Liquid Chromatography–Tandem Mass Spectrometry: Method Validation, Reference Range, and Interference Study. Diagnostics 2020, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- Succop, P.A.; Clark, S.; Chen, M.; Galke, W. Imputation of Data Values That are Less Than a Detection Limit. J. Occup. Environ. Hyg. 2004, 1, 436–441. [Google Scholar] [CrossRef]
- William, C. Handbook of Modern Hospital Safety; CRC Press: Boca Raton, FL, USA, 1999; p. 566. ISBN 9781566702560. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R. In Springer Texts in Statistics; Springer: New York, NY, USA, 2013; p. 176. ISBN 9781461471370. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Helleputte, T. LiblineaR: Linear Predictive Models Based on the Liblinear C/C++ Library R Package Version 2.10-8. 2017. Available online: ftp://ftp.us.debian.org/.1/cran/web/packages/LiblineaR/LiblineaR.pdf (accessed on 20 June 2020).
- Schliep, K.; Hechenbichler, K. kknn: Weighted k-Nearest Neighbors. R Package Version 1.3.1. 2016. Available online: https://rdrr.io/cran/kknn/ (accessed on 20 June 2020).
- Majka, M. Naivebayes: High Performance Implementation of the Naive Bayes Algorithm in R R Package Version 0.9.7; 2019. Available online: https://rdrr.io/cran/naivebayes/ (accessed on 20 June 2020).
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Soft. 2004, 11. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Soft. 2017, 77. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting R Package Version 1.0.0.2. 2020. Available online: https://cran.r-project.org/web/packages/xgboost/vignettes/xgboost.pdf (accessed on 20 June 2020).
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. JOSS 2018, 3, 861. [Google Scholar] [CrossRef]
- Konopka, T. umap: Uniform Manifold Approximation and Projection R package version 0.2.5.0. 2020. Available online: https://cran.r-project.org/web/packages/umap/index.html (accessed on 20 June 2020).
- Negrini, D.; Padoan, A.; Plebani, M. Between Web search engines and artificial intelligence: What side is shown in laboratory tests? Diagnosis 2020. [Google Scholar] [CrossRef]
- Singh, A.; Gelrud, A.; Agarwal, B. Biliary strictures: Diagnostic considerations and approach. Gastroenterol. Rep. 2015, 3, 22–31. [Google Scholar] [CrossRef]
- Bridgewater, J.; Galle, P.R.; Khan, S.A.; Llovet, J.M.; Park, J.-W.; Patel, T.; Pawlik, T.M.; Gores, G.J. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 2014, 60, 1268–1289. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In Multiple Classifier Systems; Springer: Berlin/Heidelberg, Germany, 2000; Volume 1857, pp. 1–15. ISBN 9783540677048. [Google Scholar]
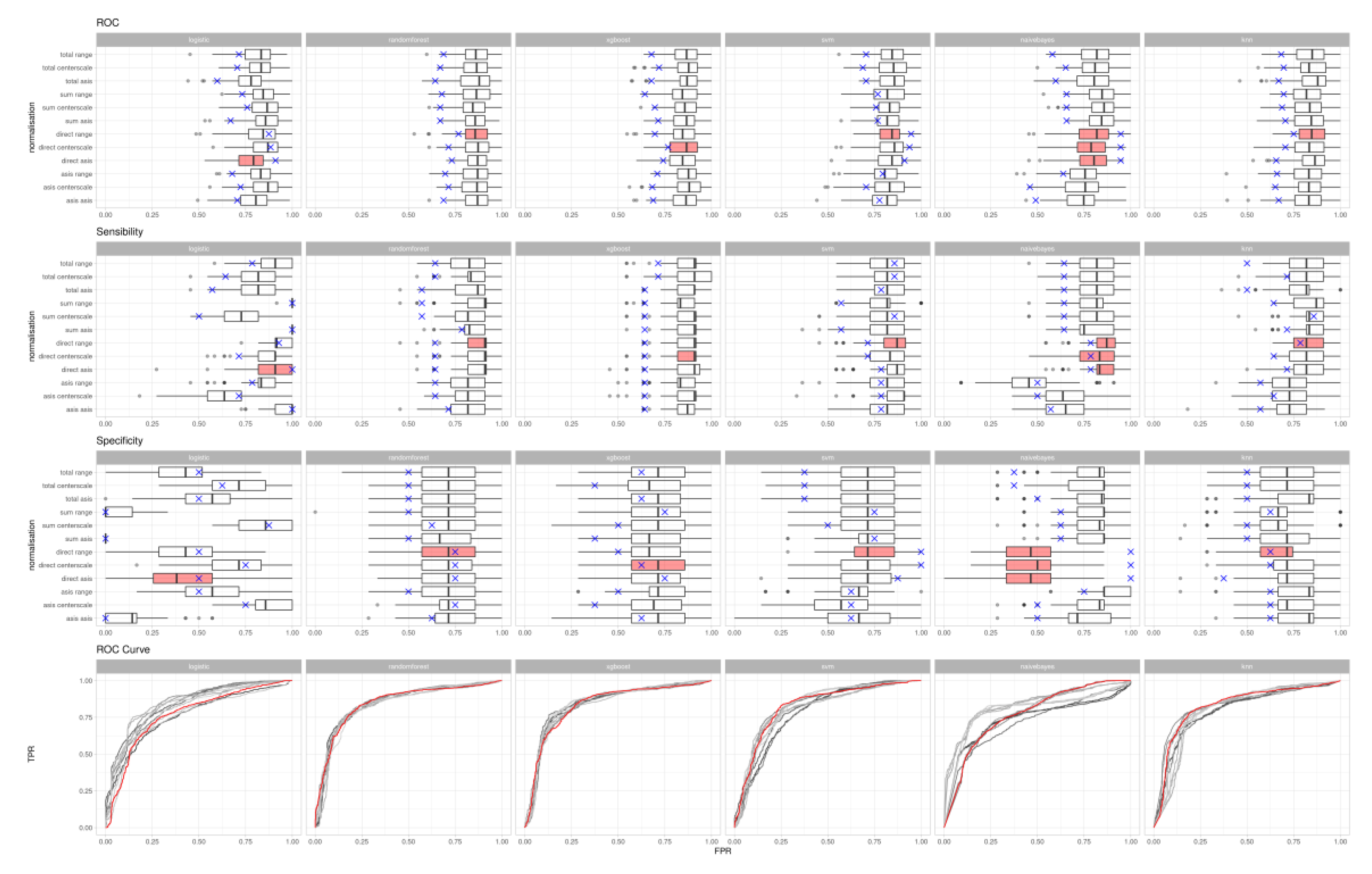

| Algorithm | Naïve Bayes | RBF Support Vector Machines | Logistic Regression | Random Forest | Extreme Gradient Boosting |
|---|---|---|---|---|---|
| First normalisation | Plasma BAs divided by the direct bilirubin | Plasma BAs divided by the direct bilirubin | Plasma BAs divided by the direct bilirubin | Plasma BAs divided by the direct bilirubin | Plasma BAs divided by the direct bilirubin |
| Second normalisation | 0–1 range | 0–1 range | No normalisation applied | 0–1 range | Mean-centring |
| True positives | 11 | 10 | 14 | 9 | 9 |
| True negatives | 8 | 8 | 4 | 6 | 5 |
| False positives | 0 | 0 | 4 | 2 | 3 |
| False Negatives | 3 | 4 | 0 | 5 | 5 |
| ROC area under curve | 0.95 | 0.95 | 0.91 | 0.77 | 0.77 |
| Accuracy | 0.864 | 0.818 | 0.818 | 0.682 | 0.700 |
| Sensitivity | 0.79 | 0.71 | 1.00 | 0.64 | 0.64 |
| Specificity | 1.00 | 1.00 | 0.50 | 0.75 | 0.63 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negrini, D.; Zecchin, P.; Ruzzenente, A.; Bagante, F.; De Nitto, S.; Gelati, M.; Salvagno, G.L.; Danese, E.; Lippi, G. Machine Learning Model Comparison in the Screening of Cholangiocarcinoma Using Plasma Bile Acids Profiles. Diagnostics 2020, 10, 551. https://doi.org/10.3390/diagnostics10080551
Negrini D, Zecchin P, Ruzzenente A, Bagante F, De Nitto S, Gelati M, Salvagno GL, Danese E, Lippi G. Machine Learning Model Comparison in the Screening of Cholangiocarcinoma Using Plasma Bile Acids Profiles. Diagnostics. 2020; 10(8):551. https://doi.org/10.3390/diagnostics10080551
Chicago/Turabian StyleNegrini, Davide, Patrick Zecchin, Andrea Ruzzenente, Fabio Bagante, Simone De Nitto, Matteo Gelati, Gian Luca Salvagno, Elisa Danese, and Giuseppe Lippi. 2020. "Machine Learning Model Comparison in the Screening of Cholangiocarcinoma Using Plasma Bile Acids Profiles" Diagnostics 10, no. 8: 551. https://doi.org/10.3390/diagnostics10080551
APA StyleNegrini, D., Zecchin, P., Ruzzenente, A., Bagante, F., De Nitto, S., Gelati, M., Salvagno, G. L., Danese, E., & Lippi, G. (2020). Machine Learning Model Comparison in the Screening of Cholangiocarcinoma Using Plasma Bile Acids Profiles. Diagnostics, 10(8), 551. https://doi.org/10.3390/diagnostics10080551










