Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal
Abstract
1. Introduction
2. Materials and Methods
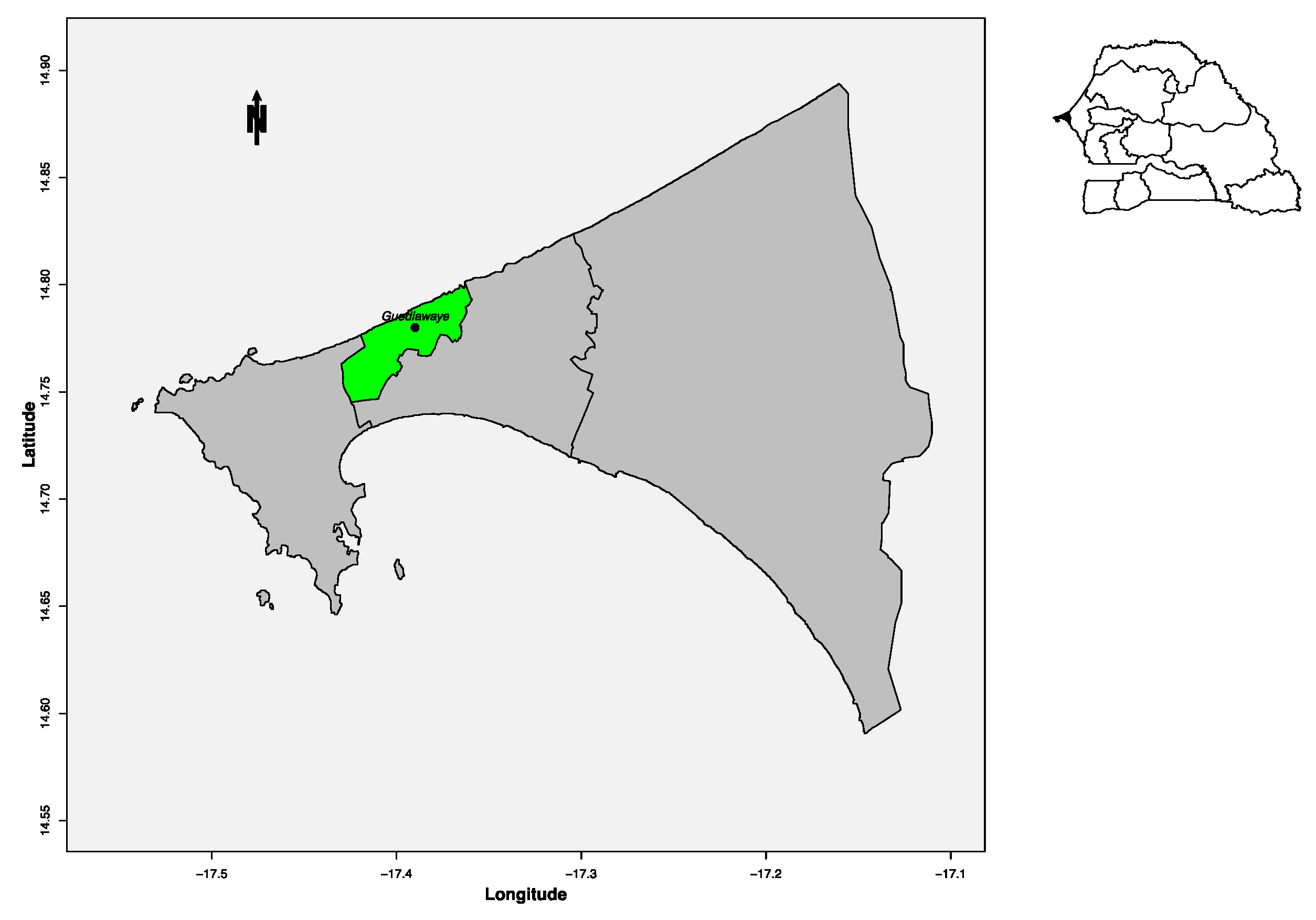
2.1. Study Site
2.2. Patient Selection
2.3. Screening Procedure in the Field
2.4. Standard Assays in the Central Laboratory
2.4.1. RNA Extraction and Real-Time qPCR
2.4.2. Non-Structural Protein 1 (NS1) Antigen Capture
2.4.3. ELISA IgM Detection
2.4.4. Viral Isolation and Immunofluorescence Indirect Assay
2.4.5. RT-PCR Amplification, Sequencing and Phylogenetic Analysis
3. Results
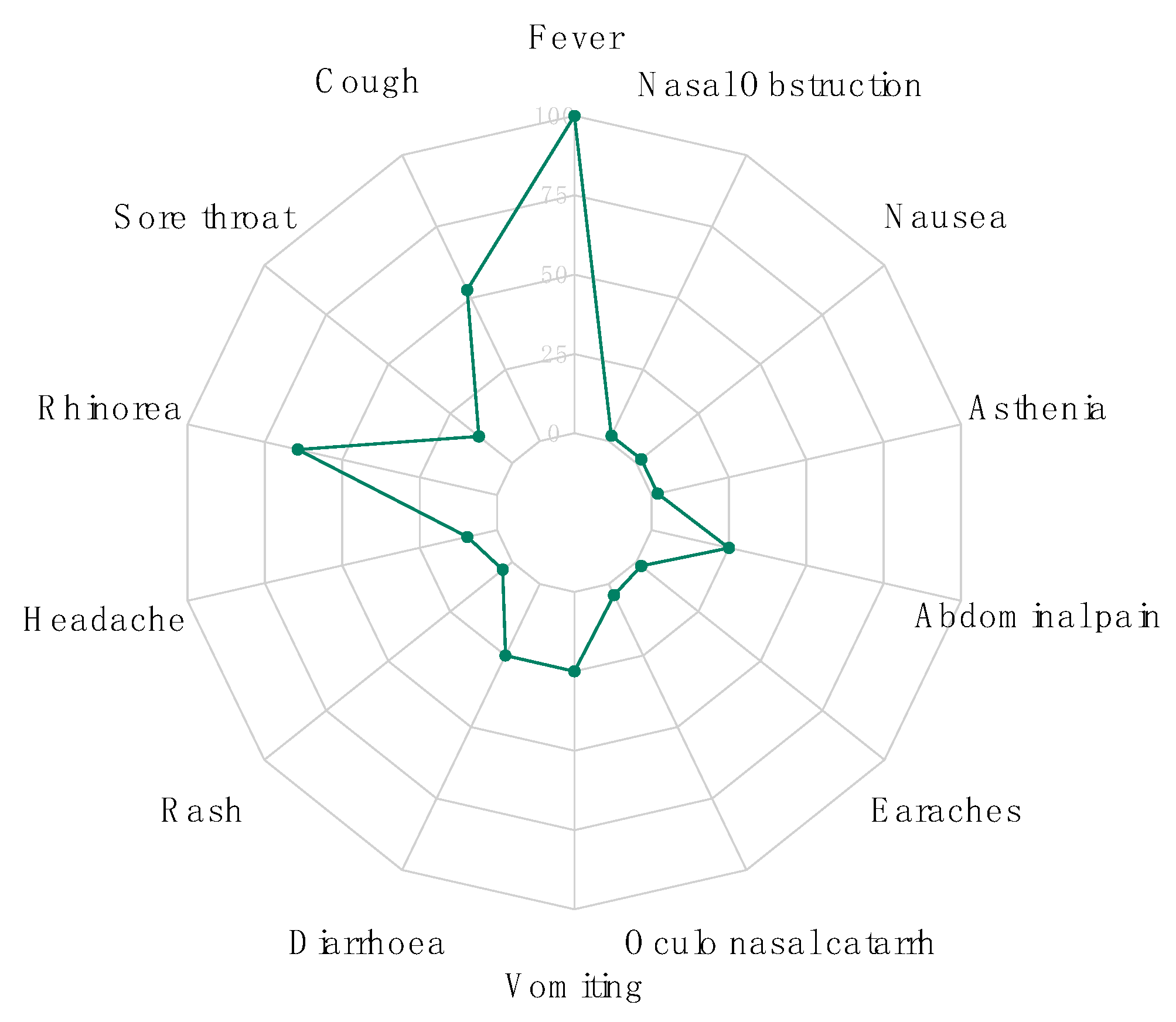
3.1. Demographic and Clinical Data
3.2. Molecular, Serologic and Antigenic Detection
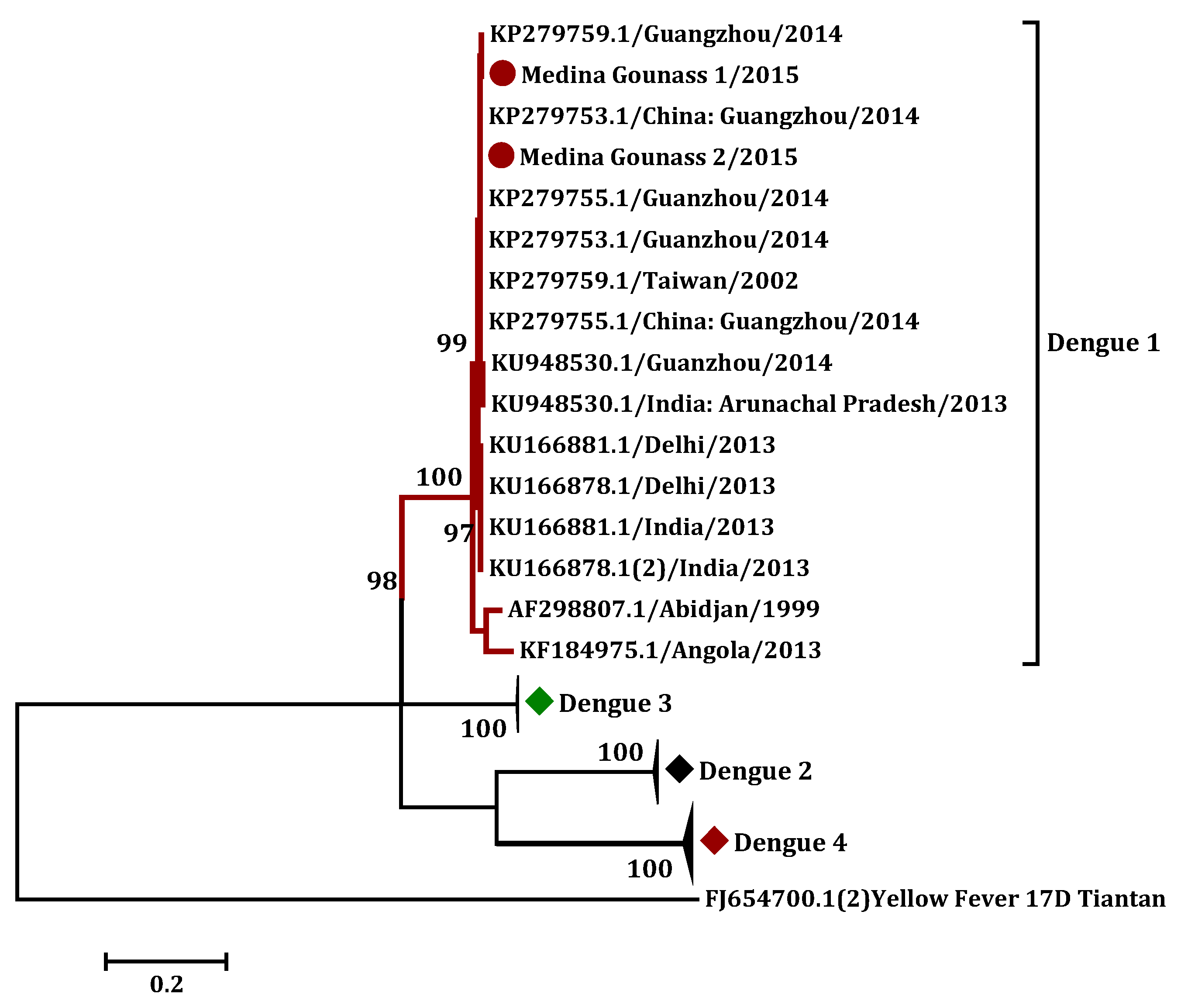
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petti, C.A.; Polage, C.R.; Quinn, T.C.; Ronald, A.R.; Sande, M.A. Laboratory Medicine in Africa: A Barrier to Effective Health Care. Clin. Infect. Dis. 2006, 42, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Feikin, D.R.; Olack, B.; Bigogo, G.M.; Audi, A.; Cosmas, L.; Aura, B.; Burke, H.; Njenga, M.K.; Williamson, J.; Breiman, R.F. The Burden of Common Infectious Disease Syndromes at the Clinic and Household Level from Population-Based Surveillance in Rural and Urban Kenya. PLoS ONE 2011, 6, e16085. [Google Scholar] [CrossRef] [PubMed]
- Okiro, E.A.; Snow, R.W. The Relationship between reported fever and Plasmodium falciparum infection in African children. Malar J. 2010, 9, 99. [Google Scholar] [CrossRef]
- WHO. World Malaria Report 2014; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Were, T.; Davenport, G.C.; Hittner, J.B.; Ouma, C.; Vulule, J.M.; Ong’Echa, J.M.; Perkins, D.J. Bacteremia in Kenyan Children Presenting with Malaria. J. Clin. Microbiol. 2010, 49, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, H. The Etiology of Febrile Illnesses among Febrile Patients Attending Felegeselam Health Center, Northwest Ethiopia. Am. J. Biomed. Life Sci. 2013, 1, 58. [Google Scholar] [CrossRef][Green Version]
- Crump, J.A.; Ramadhani, H.O.; Morrissey, A.B.; Saganda, W.; Mwako, M.S.; Yang, L.-Y.; Chow, S.-C.; Morpeth, S.C.; Reyburn, H.; Njau, B.N.; et al. Invasive Bacterial and Fungal Infections Among Hospitalized HIV-Infected and HIV-Uninfected Adults and Adolescents in Northern Tanzania. Clin. Infect. Dis. 2011, 52, 341–348. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Boil. 2006, 4, e204. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Soropogui, B.; Patel, P.; El Wahed, A.A.; Loucoubar, C.; Fall, G.; Kiory, D.; Magassouba, N.; Keita, S.; et al. Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef]
- El Wahed, A.A.; Patel, P.; Faye, O.; Thaloengsok, S.; Heidenreich, D.; Matangkasombut, P.; Manopwisedjaroen, K.; Sakuntabhai, A.; Sall, A.A.; Hufert, F.T.; et al. Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS ONE 2015, 10, e0129682. [Google Scholar] [CrossRef]
- Weidmann, M.; Faye, O.; Faye, O.; El Wahed, A.A.; Patel, P.; Batejat, C.; Manugerra, J.C.; Adjami, A.; Niedrig, M.; Hufert, F.T.; et al. Development of Mobile Laboratory for Viral Hemorrhagic Fever Detection in Africa. J. Infect. Dis. 2018, 218, 1622–1630. [Google Scholar] [CrossRef]
- Escadafal, C.; Faye, O.; Sall, A.A.; Faye, O.; Weidmann, M.; Strohmeier, O.; Von Stetten, F.; Drexler, J.; Eberhard, M.; Niedrig, M.; et al. Rapid Molecular Assays for the Detection of Yellow Fever Virus in Low-Resource Settings. PLoS Negl. Trop. Dis. 2014, 8, e2730. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; El Wahed, A.A.; Faye, O.; Prüger, P.; Kaiser, M.; Thaloengsok, S.; Ubol, S.; Sakuntabhai, A.; Leparc-Goffart, I.; Hufert, F.T.; et al. A Field-Deployable Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of the Chikungunya Virus. PLoS Negl. Trop. Dis. 2016, 10, e0004953. [Google Scholar] [CrossRef] [PubMed]
- Euler, M.; Wang, Y.; Nentwich, O.; Piepenburg, O.; Hufert, F.T.; Weidmann, M. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. J. Clin. Virol. 2012, 54, 308–312. [Google Scholar] [CrossRef] [PubMed]
- El Wahed, A.A.; Sanabani, S.S.; Faye, O.; Pessôa, R.; Patriota, J.V.; Giorgi, R.R.; Patel, P.; Böhlken-Fascher, S.; Landt, O.; Niedrig, M.; et al. Rapid Molecular Detection of Zika Virus in Acute-Phase Urine Samples Using the Recombinase Polymerase Amplification Assay. PLoS Curr. 2017, 9. [Google Scholar] [CrossRef]
- Wagner, D.; De With, K.; Huzly, D.; Hufert, F.; Weidmann, M.; Breisinger, S.; Eppinger, S.; Kern, W.V.; Bauer, T.M. Nosocomial Acquisition of Dengue. Emerg. Infect. Dis. 2004, 10, 1872–1873. [Google Scholar] [CrossRef]
- Shu, P.-Y.; Yang, C.-F.; Su, C.-L.; Chen, C.-Y.; Chang, S.-F.; Tsai, K.-H.; Cheng, C.-H.; Huang, J.-H. Two Imported Chikungunya Cases, Taiwan. Emerg. Infect. Dis. 2008, 14, 1325–1326. [Google Scholar] [CrossRef]
- Weidmann, M.; Sanchez-Seco, M.P.; Sall, A.A.; Ly, P.O.; Thiongane, Y.; Lô, M.M.; Schley, H.; Hufert, F.T. Rapid detection of important human pathogenic Phleboviruses. J. Clin. Virol. 2008, 41, 138–142. [Google Scholar] [CrossRef]
- Weidmann, M.; Faye, O.; Faye, O.; Kranaster, R.; Marx, A.; Nunes, M.R.T.; Vasconcelos, P.F.; Hufert, F.T.; Sall, A.A. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J. Clin. Virol. 2010, 48, 187–192. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Diallo, D.; Diallo, D.; Weidmann, M.; Sall, A.A. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught Mosquitoes. Virol. J. 2013, 10, 311. [Google Scholar] [CrossRef]
- Ba, F.; Loucoubar, C.; Faye, O.; Fall, G.; Mbaye, R.N.P.N.; Sembene, M.; Diallo, M.; Balde, A.T.; Sall, A.A.; Faye, O. Retrospective analysis of febrile patients reveals unnoticed epidemic of zika fever in Dielmo, Senegal, 2000. Clin. Microbiol. Infect. Dis. 2018, 3. [Google Scholar] [CrossRef]
- Digoutte, J.; Calvo-Wilson, M.; Mondo, M.; Traore-Lamizana, M.; Adam, F. Continuous cell lines and immune ascitic fluid pools in arbovirus detection. Res. Virol. 1992, 143, 417–422. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Calisher, C.H.; Gubler, D.J.; Chang, G.J.; Vorndam, A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- L’Azou, M.; Succo, T.; Kamagaté, M.; Ouattara, A.; Gilbernair, E.; Adjogoua, E.; Luxemburger, C. Dengue: Etiology of acute febrile illness in Abidjan, Côte d’Ivoire, in 2011-2012. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Oyero, O.; Ayukekbong, J.A. High dengue NS1 antigenemia in febrile patients in Ibadan, Nigeria. Virus Res. 2014, 191, 59–61. [Google Scholar] [CrossRef]
- Libraty, D.H.; Young, P.R.; Pickering, D.; Endy, T.P.; Kalayanarooj, S.; Green, S.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; Rothman, A.L. High Circulating Levels of the Dengue Virus Nonstructural Protein NS1 Early in Dengue Illness Correlate with the Development of Dengue Hemorrhagic Fever. J. Infect. Dis. 2002, 186, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Amorim, J.H.; Alves, R.P.D.S.; Boscardin, S.B.; Ferreira, L.C.D.S. The dengue virus non-structural 1 protein: Risks and benefits. Virus Res. 2014, 181, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.M.; Sistla, S.; Dhodapkar, R.; Hamide, A.; Biswal, N.; Srinivasan, B. Evaluation of NS1 Antigen Detection for Early Diagnosis of Dengue in a Tertiary Hospital in Southern India. J. Clin. Diagn. Res. 2016, 10, DC01–DC04. [Google Scholar] [CrossRef] [PubMed]
- Pothapregrada, S. The Dilemma of Reactive NS1 Antigen Test in Dengue Fever. Indian Pediatr. 2015, 52, 906–907. [Google Scholar]
- Amarasinghe, A.; Kuritsky, J.N.; Letson, G.W.; Margolis, H.S. Dengue Virus Infection in Africa. Emerg. Infect. Dis. 2011, 17, 1349–1354. [Google Scholar] [CrossRef]
- Gehrold, S. Far from altruistic: China’s presence in Senegal. 29. Available online: https://www.kas.de/c/document_library/get_file?uuid=a2cb2655-3c82-ad66-4668-8300fdac0bff&groupId=252038 (accessed on 10 June 2020).
- Simon-Loriere, E.; Kipela, J.-M.; Fernandez-Garcia, M.D.; Diagne, M.M.; Fall, I.S.; Holmes, E.; Sall, A.A.; Faye, O.; Prot, M.; Casadémont, I.; et al. Autochthonous Japanese Encephalitis with Yellow Fever Coinfection in Africa. N. Engl. J. Med. 2017, 376, 1483–1485. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.; Hernández-Triana, L.; Banyard, A. Japanese encephalitis virus infection, diagnosis and control in domestic animals|Elsevier Enhanced Reader. Vet. Microbiol. 2017, 201, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Y.; Shi, W.; Tan, S.; Pan, Y.; Cui, S.; Zhang, Q.; Dou, X.; Lv, Y.; Li, X.; et al. The first imported case of Rift Valley fever in China reveals a genetic reassortment of different viral lineages. Emerg. Microbes Infect. 2017, 6, e4–e7. [Google Scholar] [CrossRef] [PubMed]
- Stanaway, J.D.; Shepard, N.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I.; Bedi, N.; Bensenor, I.M.; Castañeda-Orjuela, C.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef]
- Leland, D.S.; Ginocchio, C.C. Role of Cell Culture for Virus Detection in the Age of Technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]

| Sample Names | RT-qPCR | RT-RPA | NS1 Antigen Capture | |||
|---|---|---|---|---|---|---|
| Ct Value | Detection Time | Threshold Time | Detection Time | D.O | Average Detection Time | |
| Medina Gounass 1 | 29.35 | 79 min | 6 min | 6 min | 6.8477 | 2 h |
| Medina Gounass 2 | 27.59 | 75 min | 5 min | 5 min | 7.9131 | 2 h |
| Medina Gounass 3 | 26.42 | 72 min | 5 min | 5 min | 9.819 | 2 h |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dieng, I.; Hedible, B.G.; Diagne, M.M.; El Wahed, A.A.; Diagne, C.T.; Fall, C.; Richard, V.; Vray, M.; Weidmann, M.; Faye, O.; et al. Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics 2020, 10, 408. https://doi.org/10.3390/diagnostics10060408
Dieng I, Hedible BG, Diagne MM, El Wahed AA, Diagne CT, Fall C, Richard V, Vray M, Weidmann M, Faye O, et al. Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics. 2020; 10(6):408. https://doi.org/10.3390/diagnostics10060408
Chicago/Turabian StyleDieng, Idrissa, Boris Gildas Hedible, Moussa Moïse Diagne, Ahmed Abd El Wahed, Cheikh Tidiane Diagne, Cheikh Fall, Vicent Richard, Muriel Vray, Manfred Weidmann, Ousmane Faye, and et al. 2020. "Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal" Diagnostics 10, no. 6: 408. https://doi.org/10.3390/diagnostics10060408
APA StyleDieng, I., Hedible, B. G., Diagne, M. M., El Wahed, A. A., Diagne, C. T., Fall, C., Richard, V., Vray, M., Weidmann, M., Faye, O., Sall, A. A., & Faye, O. (2020). Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics, 10(6), 408. https://doi.org/10.3390/diagnostics10060408






