Increased Vascular Adhesion Protein 1 (VAP-1) Levels Are Associated with Alternative M2 Macrophage Activation and Poor Prognosis for Human Gliomas
Abstract
1. Introduction
2. Materials and Methods
2.1. AOC3 Exon Expression and DNA Methylation Datasets
2.2. Specimens
2.3. Immunohistochemistry (IHC)
2.4. Pathologic Evaluation
2.5. Statistical Analysis
3. Results
3.1. VAP-1(AOC3) Expression as a Potential Biomarker for Prognosis in Patients with Gliomas
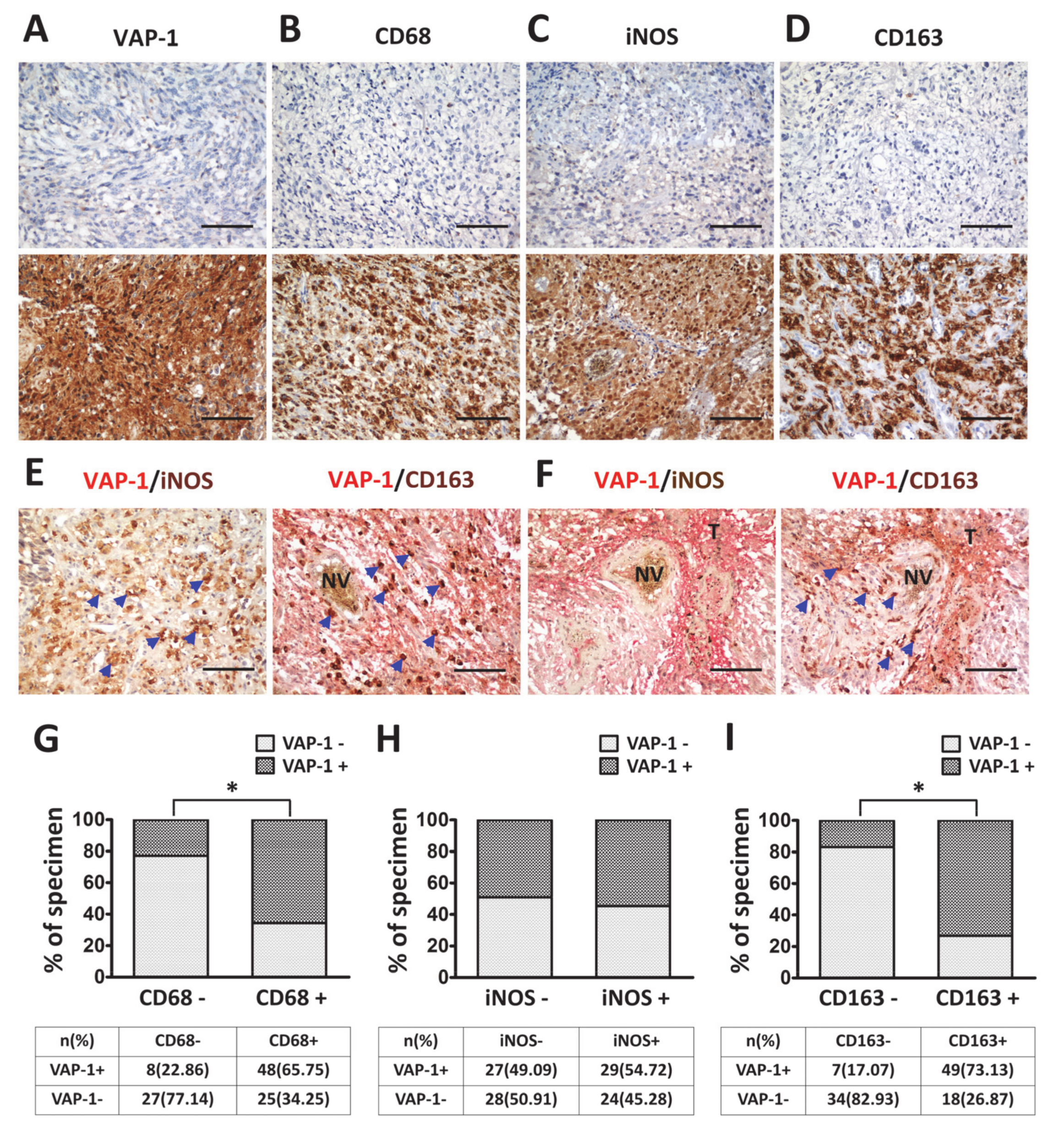
3.2. VAP-1 Expression and Co-Expressed with TAM Biomarkers are Elevated in Malignant Glioma Tissues
3.3. VAP-1 Alone and VAP-1/TAM Coexpression Correlated with Clinicopathological Variables in Glioma Patients
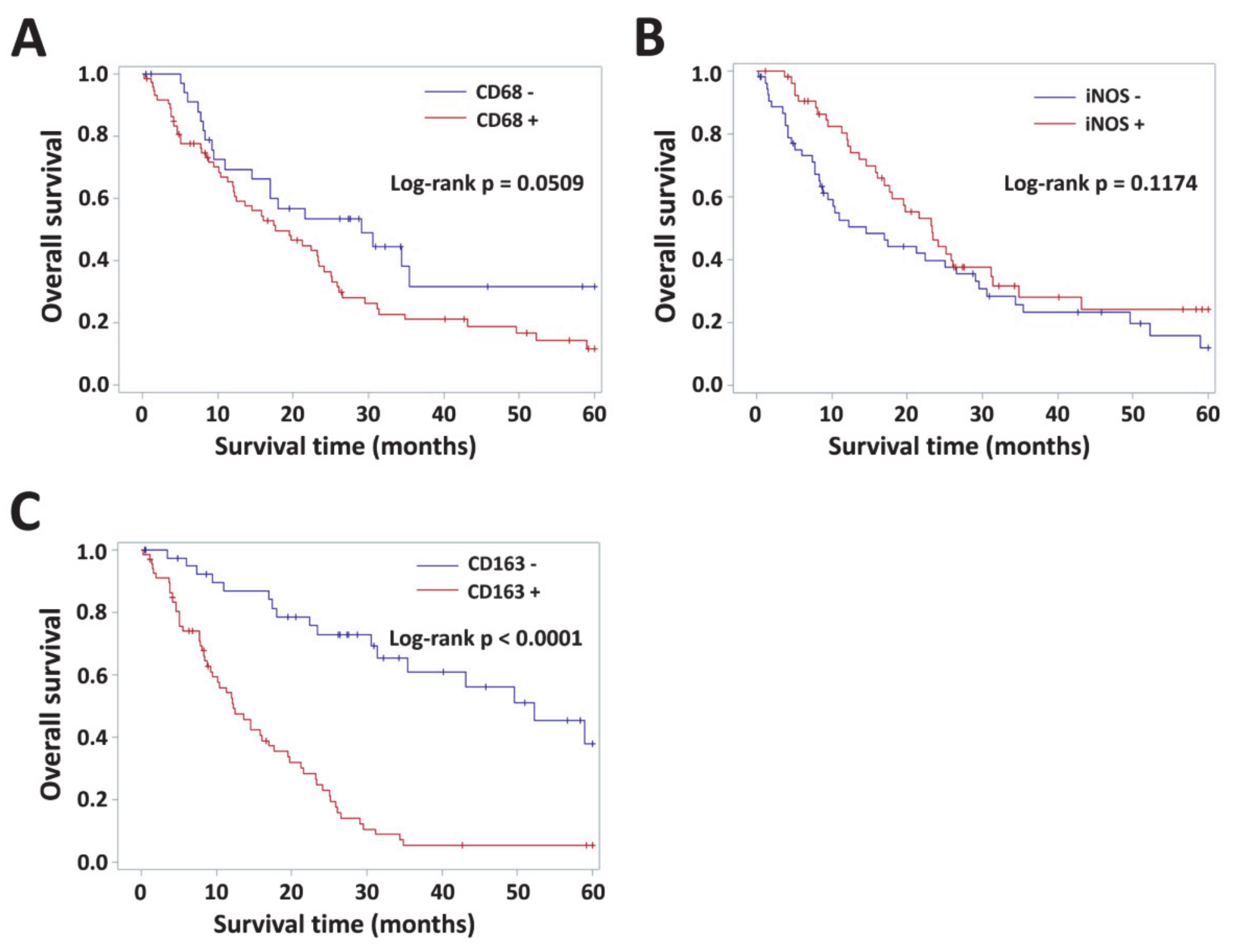
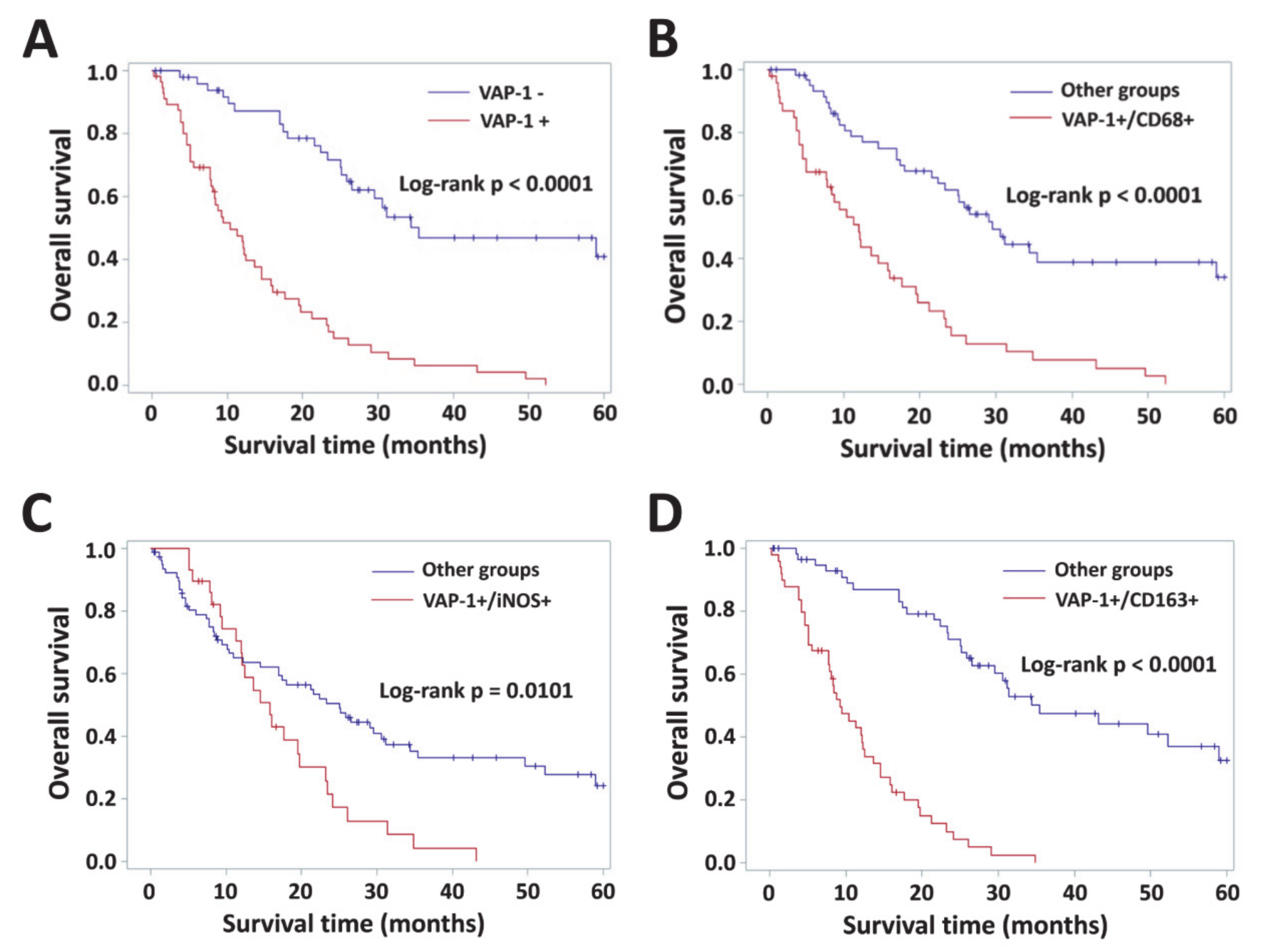
3.4. Impact of VAP-1 and VAP-1/TAM Phenotype on the Survival of Glioma Patients
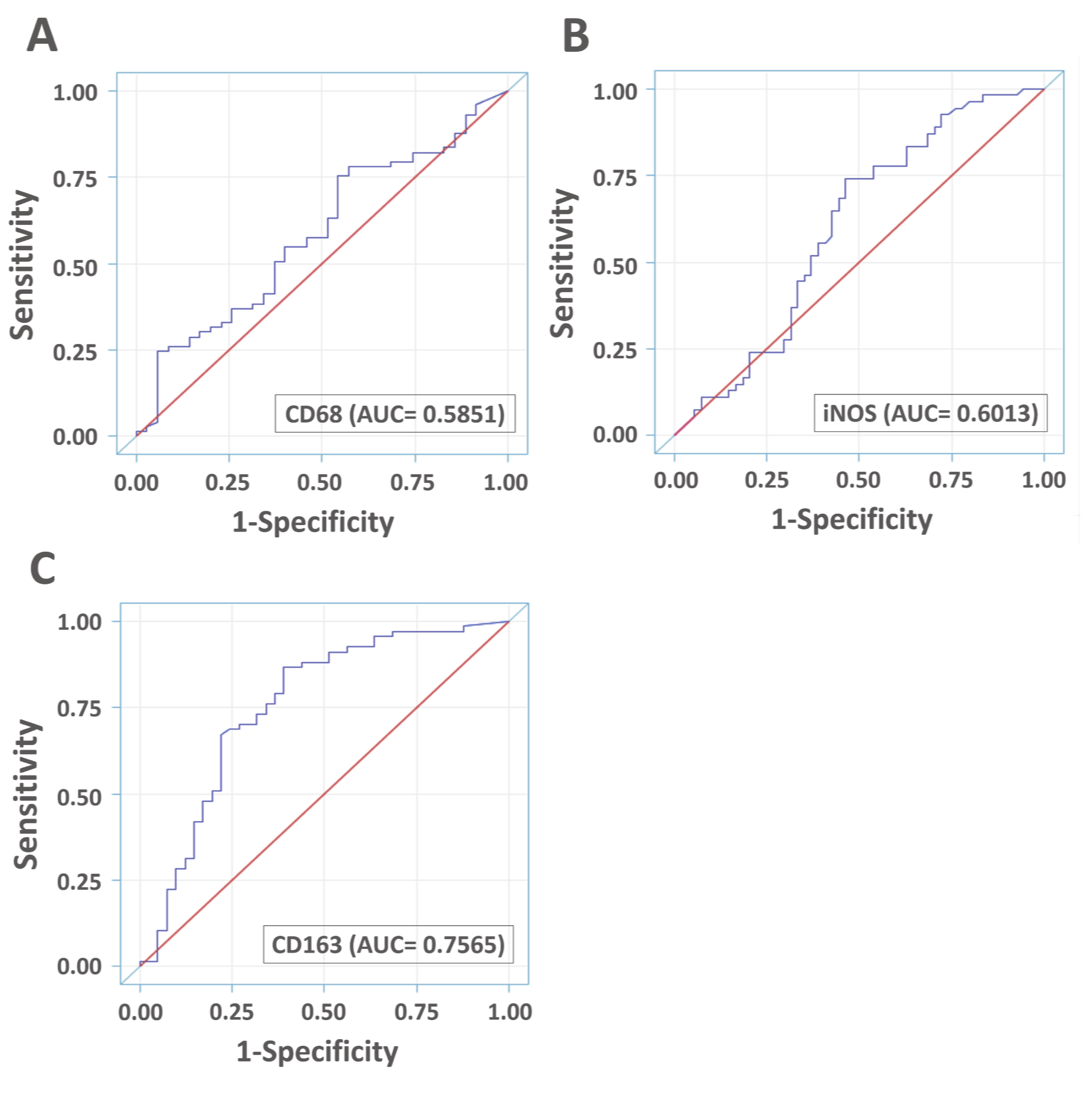
3.5. Diagnostic Accuracies of VAP-1 Expression and VAP-1/TAM Coexpression
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cheng, W.; Ren, X.; Wang, Z.; Liu, X.; Li, G.; Han, S.; Jiang, T.; Wu, A. Tumor Purity as an Underlying Key Factor in Glioma. Clin. Cancer Res. 2017, 23, 6279–6291. [Google Scholar] [CrossRef] [PubMed]
- Golebiewska, A.; Bougnaud, S.; Stieber, D.; Brons, N.H.; Vallar, L.; Hertel, F.; Klink, B.; Schrock, E.; Bjerkvig, R.; Niclou, S.P. Side population in human glioblastoma is non-tumorigenic and characterizes brain endothelial cells. Brain 2013, 136, 1462–1475. [Google Scholar] [CrossRef][Green Version]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Graeber, M.B.; Scheithauer, B.W.; Kreutzberg, G.W. Microglia in brain tumors. Glia 2002, 40, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Watters, J.J.; Schartner, J.M.; Badie, B. Microglia function in brain tumors. J. Neurosci. Res. 2005, 81, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Watabe, K. The roles of microglia/macrophages in tumor progression of brain cancer and metastatic disease. Front Biosci. (Landmark Ed) 2017, 22, 1805–1829. [Google Scholar]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Ruffell, B.; Affara, N.I.; Coussens, L.M. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012, 33, 119–126. [Google Scholar] [CrossRef]
- Lewis, C.E.; Harney, A.S.; Pollard, J.W. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 2016, 30, 18–25. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wang, N.; Li, S.; Hong, M.; Wang, X.; Feng, Y. The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxidative Med. Cell. Longev. 2016, 2016, 2795090. [Google Scholar] [CrossRef]
- Jarosz-Biej, M.; Kaminska, N.; Matuszczak, S.; Cichon, T.; Pamula-Pilat, J.; Czapla, J.; Smolarczyk, R.; Skwarzynska, D.; Kulik, K.; Szala, S. M1-like macrophages change tumor blood vessels and microenvironment in murine melanoma. PLoS ONE 2018, 13, e0191012. [Google Scholar] [CrossRef]
- Lu, G.; Zhang, R.; Geng, S.; Peng, L.; Jayaraman, P.; Chen, C.; Xu, F.; Yang, J.; Li, Q.; Zheng, H.; et al. Myeloid cell-derived inducible nitric oxide synthase suppresses M1 macrophage polarization. Nat. Commun. 2015, 6, 6676. [Google Scholar] [CrossRef] [PubMed]
- Ley, K. M1 Means Kill; M2 Means Heal. J. Immunol. 2017, 199, 2191–2193. [Google Scholar] [CrossRef]
- Kerkar, S.P.; Restifo, N.P. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012, 72, 3125–3130. [Google Scholar] [CrossRef]
- Heusinkveld, M.; van der Burg, S.H. Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 2011, 9, 216. [Google Scholar] [CrossRef]
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Mat Zin, A.A.; Ang, K.C.; Ch’ng, E.S. Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front. Oncol. 2019, 9, 1512. [Google Scholar] [CrossRef]
- Shabo, I.; Svanvik, J. Expression of macrophage antigens by tumor cells. Adv. Exp. Med. Biol. 2011, 714, 141–150. [Google Scholar] [CrossRef]
- Allavena, P.; Sica, A.; Solinas, G.; Porta, C.; Mantovani, A. The inflammatory micro-environment in tumor progression: The role of tumor-associated macrophages. Crit. Rev. Oncol./Hematol. 2008, 66, 1–9. [Google Scholar] [CrossRef]
- Roszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Komohara, Y.; Hirahara, J.; Horikawa, T.; Kawamura, K.; Kiyota, E.; Sakashita, N.; Araki, N.; Takeya, M. AM-3K, an anti-macrophage antibody, recognizes CD163, a molecule associated with an anti-inflammatory macrophage phenotype. J. Histochem. Cytochem.: Off. J. Histochem. Soc. 2006, 54, 763–771. [Google Scholar] [CrossRef]
- Sica, A.; Schioppa, T.; Mantovani, A.; Allavena, P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Potential targets of anti-cancer therapy. Eur. J. Cancer 2006, 42, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; d’Agua, B.B.; Ridley, A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 2013, 13, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Manaenko, A.; Khatibi, N.H.; Chen, W.; Zhang, J.H.; Tang, J. Vascular adhesion protein-1 inhibition provides antiinflammatory protection after an intracerebral hemorrhagic stroke in mice. J. Cereb. Blood Flow Metab. 2011, 31, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Elo, P.; Tadayon, S.; Liljenback, H.; Teuho, J.; Kakela, M.; Koskensalo, K.; Saunavaara, V.; Virta, J.; Veres, T.Z.; Kiviniemi, A.; et al. Vascular adhesion protein-1 is actively involved in the development of inflammatory lesions in rat models of multiple sclerosis. J. Neuroinflammation 2018, 15, 128. [Google Scholar] [CrossRef]
- Sole, M.; Esteban-Lopez, M.; Taltavull, B.; Fabregas, C.; Fado, R.; Casals, N.; Rodriguez-Alvarez, J.; Minano-Molina, A.J.; Unzeta, M. Blood-brain barrier dysfunction underlying Alzheimer’s disease is induced by an SSAO/VAP-1-dependent cerebrovascular activation with enhanced Abeta deposition. Biochim. Biophys Acta Mol. Basis. Dis. 2019. [Google Scholar] [CrossRef]
- Jalkanen, S.; Karikoski, M.; Mercier, N.; Koskinen, K.; Henttinen, T.; Elima, K.; Salmivirta, K.; Salmi, M. The oxidase activity of vascular adhesion protein-1 (VAP-1) induces endothelial E- and P-selectins and leukocyte binding. Blood 2007, 110, 1864–1870. [Google Scholar] [CrossRef]
- Salmi, M.; Jalkanen, S. Vascular Adhesion Protein-1: A Cell Surface Amine Oxidase in Translation. Antioxid. Redox Signal. 2017. [Google Scholar] [CrossRef]
- Salmi, M.; Jalkanen, S. Cell-surface enzymes in control of leukocyte trafficking. Nat. Rev. Immunol. 2005, 5, 760–771. [Google Scholar] [CrossRef]
- Marttila-Ichihara, F.; Auvinen, K.; Elima, K.; Jalkanen, S.; Salmi, M. Vascular adhesion protein-1 enhances tumor growth by supporting recruitment of Gr-1+CD11b+ myeloid cells into tumors. Cancer Res. 2009, 69, 7875–7883. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, H.; Luo, H.J.; Lin, Z.X.; Jiang, Z.W.; Luo, W.H. SSAO inhibitors suppress hepatocellular tumor growth in mice. Cell Immunol. 2013, 283, 61–69. [Google Scholar] [CrossRef]
- Ferjancic, S.; Gil-Bernabe, A.M.; Hill, S.A.; Allen, P.D.; Richardson, P.; Sparey, T.; Savory, E.; McGuffog, J.; Muschel, R.J. VCAM-1 and VAP-1 recruit myeloid cells that promote pulmonary metastasis in mice. Blood 2013, 121, 3289–3297. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Mazzieri, R.; Yang, T.; Gobe, G.C. Translational Significance for Tumor Metastasis of Tumor-Associated Macrophages and Epithelial-Mesenchymal Transition. Front. Immunol. 2017, 8, 1106. [Google Scholar] [CrossRef] [PubMed]
- Mukaida, N.; Nosaka, T.; Nakamoto, Y.; Baba, T. Lung Macrophages: Multifunctional Regulator Cells for Metastatic Cells. Int. J. Mol. Sci. 2018. [Google Scholar] [CrossRef]
- Nakao, S.; Noda, K.; Zandi, S.; Sun, D.; Taher, M.; Schering, A.; Xie, F.; Mashima, Y.; Hafezi-Moghadam, A. VAP-1-mediated M2 macrophage infiltration underlies IL-1beta- but not VEGF-A-induced lymph- and angiogenesis. Am. J. Pathol. 2011, 178, 1913–1921. [Google Scholar] [CrossRef]
- Kostoro, J.; Chang, S.J.; Clark Lai, Y.C.; Wu, C.C.; Chai, C.Y.; Kwan, A.L. Overexpression of vascular adhesion protein-1 is associated with poor prognosis of astrocytomas. APMIS 2016, 124, 462–468. [Google Scholar] [CrossRef]
- Fukuhara, J.; Kase, S.; Noda, K.; Murata, M.; Noda, M.; Ando, R.; Dong, Z.; Kanda, A.; Ishida, S. Immunolocalization of vascular adhesion protein-1 in human conjunctival tumors. Ophthalmic Res. 2012, 48, 33–37. [Google Scholar] [CrossRef]
- Li, Y.I.; Hung, J.S.; Yu, T.Y.; Liou, J.M.; Wei, J.N.; Kao, H.L.; Chuang, L.M.; Shun, C.T.; Lee, P.H.; Lai, H.S.; et al. Serum vascular adhesion protein-1 predicts all-cause mortality and cancer-related mortality in subjects with colorectal cancer. Clin. Chim. Acta 2014, 428, 51–56. [Google Scholar] [CrossRef]
- Lai, Y.C.; Chang, S.J.; Kostoro, J.; Kwan, A.L.; Chai, C.Y. Vascular adhesion protein-1 as indicator of breast cancer tumor aggressiveness and invasiveness. APMIS 2018, 126, 755–761. [Google Scholar] [CrossRef]
- Varis, A.; Wolf, M.; Monni, O.; Vakkari, M.L.; Kokkola, A.; Moskaluk, C.; Frierson, H., Jr.; Powell, S.M.; Knuutila, S.; Kallioniemi, A.; et al. Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res. 2002, 62, 2625–2629. [Google Scholar] [PubMed]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der. Pathol. 1987, 8, 138–140. [Google Scholar]
- Fedchenko, N.; Reifenrath, J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue - a review. Diagnostic Pathol. 2014, 9, 221. [Google Scholar] [CrossRef]
- Marttila-Ichihara, F.; Castermans, K.; Auvinen, K.; Oude Egbrink, M.G.; Jalkanen, S.; Griffioen, A.W.; Salmi, M. Small-molecule inhibitors of vascular adhesion protein-1 reduce the accumulation of myeloid cells into tumors and attenuate tumor growth in mice. J. Immunol. 2010, 184, 3164–3173. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Ohnishi, K.; Kuratsu, J.; Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008, 216, 15–24. [Google Scholar] [CrossRef]
- Annovazzi, L.; Mellai, M.; Bovio, E.; Mazzetti, S.; Pollo, B.; Schiffer, D. Microglia immunophenotyping in gliomas. Oncol. Lett. 2018, 15, 998–1006. [Google Scholar] [CrossRef]
- Kennedy, B.C.; Showers, C.R.; Anderson, D.E.; Anderson, L.; Canoll, P.; Bruce, J.N.; Anderson, R.C. Tumor-associated macrophages in glioma: Friend or foe? J. Oncol. 2013, 2013, 486912. [Google Scholar] [CrossRef]
- Gabrusiewicz, K.; Rodriguez, B.; Wei, J.; Hashimoto, Y.; Healy, L.M.; Maiti, S.N.; Thomas, G.; Zhou, S.; Wang, Q.; Elakkad, A.; et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016. [Google Scholar] [CrossRef]
- Gutmann, D.H.; Kettenmann, H. Microglia/Brain Macrophages as Central Drivers of Brain Tumor Pathobiology. Neuron 2019, 104, 442–449. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Jetten, N.; Verbruggen, S.; Gijbels, M.J.; Post, M.J.; De Winther, M.P.; Donners, M.M. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 2014, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Corliss, B.A.; Azimi, M.S.; Munson, J.M.; Peirce, S.M.; Murfee, W.L. Macrophages: An Inflammatory Link Between Angiogenesis and Lymphangiogenesis. Microcirculation 2016, 23, 95–121. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, C.; Muthana, M.; Coffelt, S.B.; Lewis, C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer 2008, 8, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Larghi, P.; Mancino, A.; Rubino, L.; Porta, C.; Totaro, M.G.; Rimoldi, M.; Biswas, S.K.; Allavena, P.; Mantovani, A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008, 18, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]


| Characteristics | Median (Range) or n (%) |
|---|---|
| Total number of patients | 108 |
| Age (years) mean ± SD medians (corresponding ranges) | 50.26 ± 17.51 52.00 (20.00–83.00) |
| Tumor size (cm) mean ± SD medians (corresponding ranges) | 1.88 ± 1.32 1.60(0.20–7.70) |
| Follow-up of the patient cohort (months) mean ± SD medians (corresponding ranges) | 20.88 ± 17.39 16.82 (0.23–60.00) |
| Gender, n (%) Female Male | 40 (37.04) 68 (62.96) |
| WHO grade, n (%) II III IV | 27 (25.00) 35 (32.41) 46 (42.59) |
| Histological type, n (%) diffuse astrocytoma oligoastrocytoma oligodendroglioma anaplstic astrocytoma GBM | 28 (25.93) 10 (9.26) 3 (2.78) 28 (25.93) 39 (36.11) |
| Recurrence, n (%) Absent Present | 53 (49.07) 55 (50.93) |
| Survival status, n (%) survived died | 34 (31.48) 74 (68.52) |
| IDH1 mutant, n (%) Negative Positive | 81 (75.00) 27 (25.00) |
| Parameters | VAP-1- n (%) | VAP-1+ n (%) | p Value | Other Groups * n (%) | VAP-1+/ CD68+ n (%) | p Value | Other Groups ** n (%) | VAP-1+/ iNOS+ n (%) | p Value | Other Groups *** n (%) | VAP-1+/ CD163+ n (%) | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 52 (48.15) | 56 (51.85) | 61 (56.48) | 47 (43.52) | 79 (73.15) | 29 (26.85) | 59 (54.63) | 49 (45.37) | |||||
| Gender Female Male | 21 (40.38) 31 (59.62) | 19 (33.93) 37 (66.07) | 0.4876 | 25 (40.98) 36 (59.02) | 15(31.91) 32(68.09) | 0.3333 | 31 (39.24) 48 (60.76) | 9 (31.03) 20 (68.97) | 0.4338 | 24 (40.68) 35 (59.32) | 16 (32.65) 33 (67.35) | 0.3899 |
| Age ≤45 years >45 years | 25 (48.08) 27 (51.92) | 17 (30.36) 39 (69.64) | 0.0591 | 28 (45.90) 33 (54.10) | 14 (29.79) 33 (70.21) | 0.0885 | 33 (41.77) 46 (58.23) | 9 (31.03) 20 (68.97) | 0.3104 | 29 (49.15) 30 (50.85) | 13 (26.53) 36 (73.47) | 0.0164 |
| WHO grade II III IV | 24 (46.15) 21 (40.38) 7 (13.46) | 3 (5.36) 14 (25.00) 39 (69.64) | <0.0001 | 24 (39.34) 22 (36.07) 15 (24.59) | 3 (6.38) 13 (27.66) 31 (65.96) | <0.0001 | 26 (32.91) 31 (39.24) 22 (27.85) | 1 (3.45) 4 (13.79) 24 (82.76) | <0.0001 | 25 (42.37) 24 (40.68) 10 (16.95) | 2 (4.08) 11 (22.45) 36 (73.47) | <0.0001 |
| Tumor size <2 cm ≥2 cm | 33 (63.46) 19 (36.54) | 30 (53.57) 26 (46.43) | 0.2976 | 39 (63.93) 22 (36.07) | 24 (51.06) 23 (48.94) | 0.1786 | 47 (59.49) 32 (40.51) | 16 (55.17) 13 (44.83) | 0.6864 | 38 (64.41) 21 (35.59) | 25 (51.02) 24 (48.98) | 0.1601 |
| Recurrence Absent Present | 25 (48.08) 27 (51.92) | 28 (50.00) 28 (50.00) | 0.8417 | 30 (49.18) 31 (50.82) | 23 (48.94) 24 (51.06) | 0.9799 | 41 (51.90) 38 (48.10) | 12 (41.38) 17 (58.62) | 0.3325 | 29 (49.15) 30 (50.85) | 24 (48.98) 25 (51.02) | 0.9857 |
| Survival status survived died | 29 (55.77) 23 (44.23) | 5 (8.93) 51 (91.07) | <0.0001 | 29 (47.54) 32 (52.46) | 5 (10.64) 42 (89.36) | <0.0001 | 30 (37.97) 49 (62.03) | 4 (13.79) 25 (86.21) | 0.0165 | 30 (50.85) 29 (49.15) | 4 (8.16) 45 (91.84) | <0.0001 |
| IDH1 mutant Negative Positive | 34 (65.38) 18 (34.62) | 47 (83.93) 9 (16.07) | 0.0262 | 43 (70.49) 18 (29.51) | 38 (80.85) 9 (19.15) | 0.2177 | 57 (72.15) 22 (27.85) | 24 (82.76) 5 (17.24) | 0.2592 | 38 (64.41) 21 (35.59) | 43 (87.76) 6 (12.24) | 0.0053 |
| Variable | VAP-1 | CD68 | iNOS | CD163 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Correlation | p Value | Correlation | p Value | Correlation | p Value | Correlation | p Value | ||
| VAP-1 | 1.00000 | – | 0.40181 | <0.0001 | 0.05629 | 0.5628 | 0.54450 | <0.0001 | |
| CD68 | 0.40181 | <0.0001 | 1.00000 | – | 0.04654 | 0.6325 | 0.35519 | 0.0002 | |
| iNOS | 0.05629 | 0.5628 | 0.04654 | 0.6325 | 1.00000 | – | 0.11909 | 0.2196 | |
| CD163 | 0.54450 | <0.0001 | 0.35519 | 0.0002 | 0.11909 | 0.2196 | 1.00000 | – | |
| Parameters | Univariate | Multivariate | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p-Value | VAP-1 | p-Value | VAP-1/CD68 | p-Value | VAP-1/iNOS | p-Value | VAP-1/CD163 | p-Value | |
| HR (95% CI) | HR (95% CI) | HR (95% CI) | HR (95% CI) | |||||||
| VAP-1 | 5.057 (3.016–8.481) | <0.0001 | 4.688(2.736–8.033) | <0.0001 | – | – | – | – | – | – |
| VAP-1/CD68 | 3.483 (2.164–5.605) | <0.0001 | – | – | 3.226 (1.980–5.256) | <0.0001 | – | – | – | – |
| VAP-1/iNOS | 1.908 (1.157–3.145) | 0.0113 | – | – | – | – | 1.572 (0.945–2.615) | 0.0817 | – | – |
| VAP-1/CD163 | 7.047 (4.085–12.155) | <0.0001 | – | – | – | – | 6.597 (3.677–11.836) | <0.0001 | ||
| Gender (female = 1) | 0.956 (0.597–1.531) | 0.8513 | 0.735 (0.426–1.267) | 0.2675 | 0.725 (0.419–1.252) | 0.2480 | 0.736 (0.436–1.240) | 0.2495 | 0.652 (0.372–1.140) | 0.1336 |
| Age | 1.853 (1.140–3.012) | 0.0129 | 1.816 (1.071–3.077) | 0.0266 | 1.886 (1.117–3.185) | 0.0176 | 1.817 (1.090–3.028) | 0.0219 | 1.755 (1.014–3.039) | 0.0445 |
| Tumor size | 1.231 (0.776–1.953) | 0.3765 | 1.645 (0.955–2.835) | 0.0730 | 1.474 (0.866–2.510) | 0.1526 | 1.411 (0.849–2.347) | 0.1843 | 1.438 (0.848–2.439) | 0.1774 |
| Recurrence | 0.924 (0.583–1.465) | 0.7368 | 0.848 (0.516–1.392) | 0.5144 | 0.900 (0.552–1.465) | 0.6709 | 1.043 (0.648–1.679) | 0.8619 | 0.782 (0.470–1.304) | 0.3464 |
| IDH1 mutant | 0.356 (0.191–0.662) | 0.0011 | 0.517 (0.270–0.993) | 0.0474 | 0.455 (0.239–0.866) | 0.0165 | 0.412 (0.216–0.782) | 0.0067 | 0.533 (0.279–1.019) | 0.0569 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, S.-J.; Tu, H.-P.; Lai, Y.-C.C.; Luo, C.-W.; Nejo, T.; Tanaka, S.; Chai, C.-Y.; Kwan, A.-L. Increased Vascular Adhesion Protein 1 (VAP-1) Levels Are Associated with Alternative M2 Macrophage Activation and Poor Prognosis for Human Gliomas. Diagnostics 2020, 10, 256. https://doi.org/10.3390/diagnostics10050256
Chang S-J, Tu H-P, Lai Y-CC, Luo C-W, Nejo T, Tanaka S, Chai C-Y, Kwan A-L. Increased Vascular Adhesion Protein 1 (VAP-1) Levels Are Associated with Alternative M2 Macrophage Activation and Poor Prognosis for Human Gliomas. Diagnostics. 2020; 10(5):256. https://doi.org/10.3390/diagnostics10050256
Chicago/Turabian StyleChang, Shu-Jyuan, Hung-Pin Tu, Yen-Chang Clark Lai, Chi-Wen Luo, Takahide Nejo, Shota Tanaka, Chee-Yin Chai, and Aij-Lie Kwan. 2020. "Increased Vascular Adhesion Protein 1 (VAP-1) Levels Are Associated with Alternative M2 Macrophage Activation and Poor Prognosis for Human Gliomas" Diagnostics 10, no. 5: 256. https://doi.org/10.3390/diagnostics10050256
APA StyleChang, S.-J., Tu, H.-P., Lai, Y.-C. C., Luo, C.-W., Nejo, T., Tanaka, S., Chai, C.-Y., & Kwan, A.-L. (2020). Increased Vascular Adhesion Protein 1 (VAP-1) Levels Are Associated with Alternative M2 Macrophage Activation and Poor Prognosis for Human Gliomas. Diagnostics, 10(5), 256. https://doi.org/10.3390/diagnostics10050256





