MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Patient Selection
2.3. Baseline Measurements
2.4. PAD Assessment and Sample Processing
2.5. miRNA Isolation, Quantification and Purity Analysis
2.6. miRNA Bioinformatics Analysis
2.7. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.8. Target Prediction, Visualization and Over-Representation Analysis
2.9. Statistical Methods
3. Results
3.1. Cohort Description
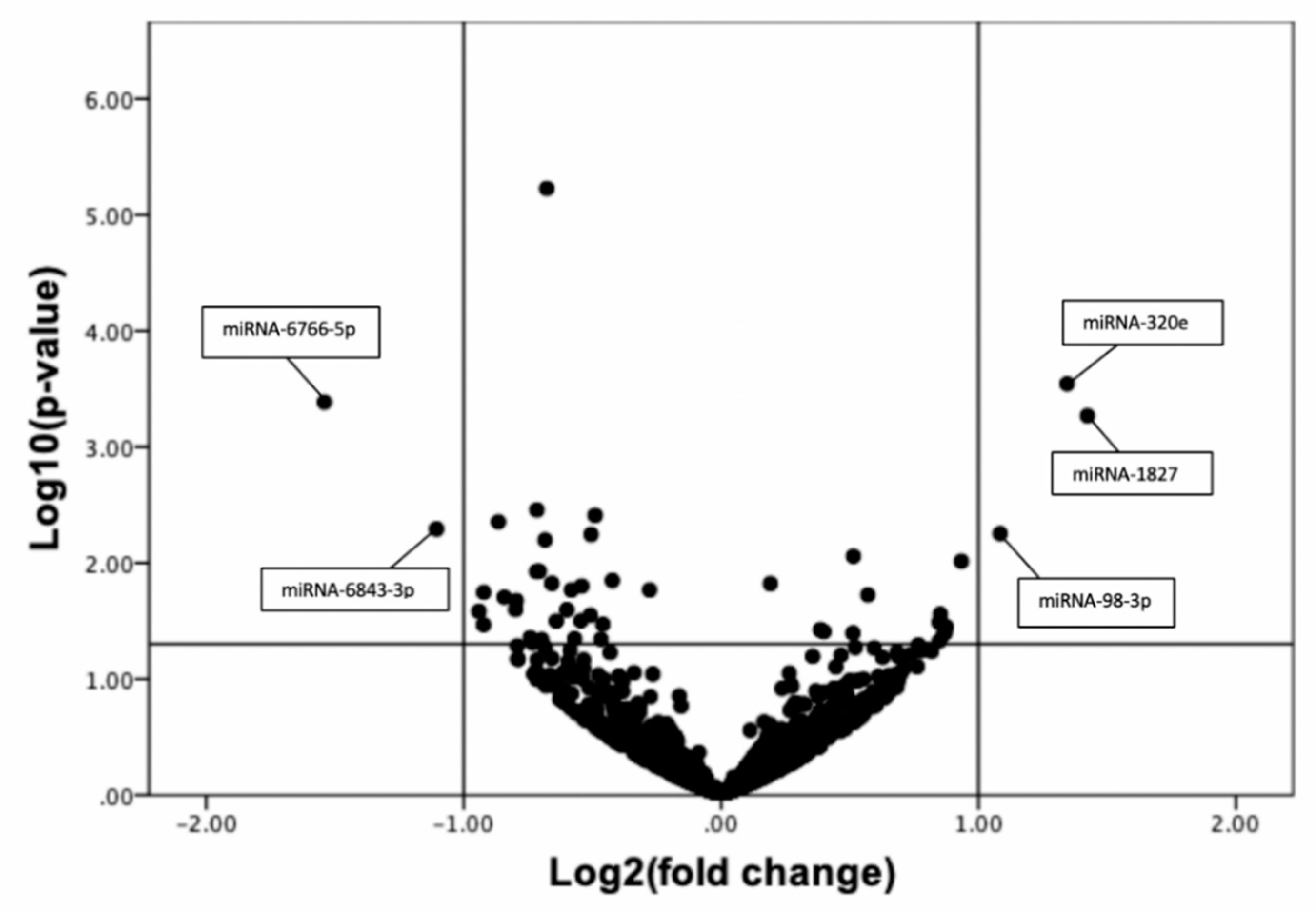
3.2. Discovery Phase: Global Plasma miRNA Profile of CLTI and Non-PAD Patients Using Next Generation Sequencing
3.3. Confirmation of Next Generation Sequencing Results by Quantitative qRT-PCR
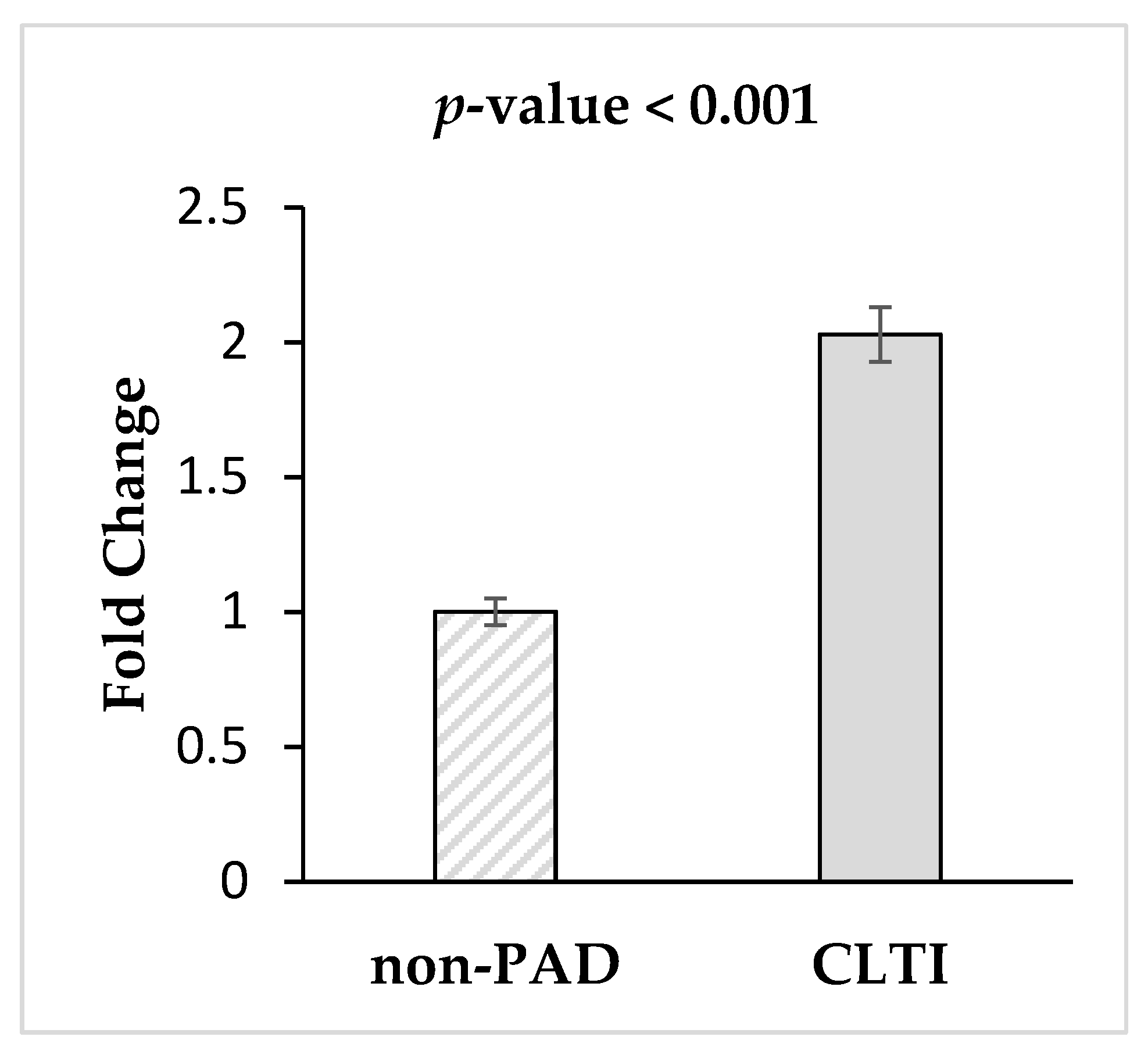
3.4. Validation of miRNA-1827via qRT-PCR
3.5. miRNA-1827 Target Prediction, Function and Pathway Analysis.
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Fowkes, F.G.; Rudan, D.; Rudan, I.; Aboyans, V.; Denenberg, J.O.; McDermott, M.M.; Norman, P.E.; Sampson, U.K.; Williams, L.J.; Mensah, G.A.; et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: A systematic review and analysis. Lancet 2013, 382, 1329–1340. [Google Scholar] [CrossRef]
- Bonow, R.O.; Smaha, L.A.; Smith, S.C., Jr.; Mensah, G.A.; Lenfant, C. World Heart Day 2002: The international burden of cardiovascular disease: Responding to the emerging global epidemic. Circulation 2002, 106, 1602–1605. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Santulli, G. Update on peripheral artery disease: Epidemiology and evidence-based facts. Atherosclerosis 2018, 275, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Hankey, G.J.; Norman, P.E.; Eikelboom, J.W. Medical treatment of peripheral arterial disease. JAMA 2006, 295, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Peach, G.; Griffin, M.; Jones, K.G.; Thompson, M.M.; Hinchliffe, R.J. Diagnosis and management of peripheral arterial disease. BMJ 2012, 345, e5208. [Google Scholar] [CrossRef] [PubMed]
- Clair, D.; Shah, S.; Weber, J. Current state of diagnosis and management of critical limb ischemia. Curr. Cardiol. Rep. 2012, 14, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Varu, V.N.; Hogg, M.E.; Kibbe, M.R. Critical limb ischemia. J. Vasc. Surg. 2010, 51, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Norgren, L.; Hiatt, W.R.; Dormandy, J.A.; Nehler, M.R.; Harris, K.A.; Fowkes, F.G.; Group, T.I.W. Inter-Society Consensus for the Management of Peripheral Arterial Disease (TASC II). J. Vasc. Surg. 2007, 45 (Suppl. S), S5–S67. [Google Scholar] [CrossRef]
- Santilli, J.D.; Santilli, S.M. Chronic critical limb ischemia: Diagnosis, treatment and prognosis. Am. Fam. Physician 1999, 59, 1899–1908. [Google Scholar]
- Ohtake, T.; Oka, M.; Ikee, R.; Mochida, Y.; Ishioka, K.; Moriya, H.; Hidaka, S.; Kobayashi, S. Impact of lower limbs’ arterial calcification on the prevalence and severity of PAD in patients on hemodialysis. J. Vasc. Surg. 2011, 53, 676–683. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Kim, Y.S. Peripheral Arterial Disease in Patients with Type 2 Diabetes Mellitus. Diabetes Metab. J. 2015, 39, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Mayr, M. MicroRNAs in vascular and metabolic disease. Circ. Res. 2012, 110, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of microRNAs: A review. J. Physiol. Biochem. 2011, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Pereira-da-Silva, T.; Coutinho Cruz, M.; Carrusca, C.; Cruz Ferreira, R.; Napoleao, P.; Mota Carmo, M. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: A systematic review. Am. J. Cardiovasc. Dis. 2018, 8, 1–13. [Google Scholar]
- Stather, P.W.; Sylvius, N.; Wild, J.B.; Choke, E.; Sayers, R.D.; Bown, M.J. Differential microRNA expression profiles in peripheral arterial disease. Circ. Cardiovasc. Genet. 2013, 6, 490–497. [Google Scholar] [CrossRef]
- Signorelli, S.S.; Volsi, G.L.; Pitruzzella, A.; Fiore, V.; Mangiafico, M.; Vanella, L.; Parenti, R.; Rizzo, M.; Volti, G.L. Circulating miR-130a, miR-27b, and miR-210 in Patients With Peripheral Artery Disease and Their Potential Relationship With Oxidative Stress. Angiology 2016, 67, 945–950. [Google Scholar] [CrossRef]
- Lavrik, I.N.; Golks, A.; Krammer, P.H. Caspases: Pharmacological manipulation of cell death. J. Clin. Investig. 2005, 115, 2665–2672. [Google Scholar] [CrossRef]
- Siddiqui, W.A.; Ahad, A.; Ahsan, H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: An update. Arch. Toxicol. 2015, 89, 289–317. [Google Scholar] [CrossRef]
- van Rooij, E.; Olson, E.N. MicroRNA therapeutics for cardiovascular disease: Opportunities and obstacles. Nat. Rev. Drug. Discov. 2012, 11, 860–872. [Google Scholar] [CrossRef]
- Small, E.M.; Frost, R.J.; Olson, E.N. MicroRNAs add a new dimension to cardiovascular disease. Circulation 2010, 121, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci. Transl. Med. 2014, 6, 239ps233. [Google Scholar] [CrossRef] [PubMed]
- Annex, B.H. Is a simple biomarker for peripheral arterial disease on the horizon? Am. Heart Assoc. 2007, 116, 1346–1348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qadura, M.; Terenzi, D.C.; Verma, S.; Al-Omran, M.; Hess, D.A. Concise review: Cell therapy for critical limb ischemia: An integrated review of preclinical and clinical studies. Stem Cells 2018, 36, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Zamzam, A.; Syed, M.H.; Rand, M.L.; Singh, K.; Hussain, M.A.; Jain, S.; Khan, H.; Verma, S.; Al-Omran, M.; Abdin, R.; et al. Altered coagulation profile in peripheral artery disease patients. Vascular 2020. [Google Scholar] [CrossRef]
- Li, J.Y.; Cheng, B.; Wang, X.F.; Wang, Z.J.; Zhang, H.M.; Liu, S.F.; Chen, L.S.; Huang, W.J.; Liu, J.; Deng, A.P. Circulating MicroRNA-4739 May Be a Potential Biomarker of Critical Limb Ischemia in Patients with Diabetes. Biomed. Res. Int. 2018, 2018, 4232794. [Google Scholar] [CrossRef]
- Cheng, B.; Li, J.-y.; Li, X.-c.; Wang, X.-f.; Wang, Z.-j.; Liu, J.; Deng, A.-p. MiR-323b-5p acts as a novel diagnostic biomarker for critical limb ischemia in type 2 diabetic patients. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Desjarlais, M.; Dussault, S.; Dhahri, W.; Mathieu, R.; Rivard, A. MicroRNA-150 Modulates Ischemia-Induced Neovascularization in Atherosclerotic Conditions. Arter. Thromb. Vasc. Biol. 2017, 37, 900–908. [Google Scholar] [CrossRef]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef]
- Grundmann, S.; Hans, F.P.; Kinniry, S.; Heinke, J.; Helbing, T.; Bluhm, F.; Sluijter, J.P.; Hoefer, I.; Pasterkamp, G.; Bode, C.; et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 2011, 123, 999–1009. [Google Scholar] [CrossRef]
- Semo, J.; Sharir, R.; Afek, A.; Avivi, C.; Barshack, I.; Maysel-Auslender, S.; Krelin, Y.; Kain, D.; Entin-Meer, M.; Keren, G.; et al. The 106b approximately 25 microRNA cluster is essential for neovascularization after hindlimb ischaemia in mice. Eur. Heart J. 2014, 35, 3212–3223. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Noncoding RNAs in Critical Limb Ischemia. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 523–533. [Google Scholar] [CrossRef]
- Yin, K.-J.; Olsen, K.; Hamblin, M.; Zhang, J.; Schwendeman, S.P.; Chen, Y.E. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J. Biol. Chem. 2012, 287, 27055–27064. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Ren, S.; Lerchenmüller, C.; Sun, J.; Weiss, N.; Most, P.; Peppel, K. MicroRNA-138 regulates hypoxia-induced endothelial cell dysfunction by targeting S100A1. PLoS ONE 2013, 8, e78684. [Google Scholar] [CrossRef]
- Biswas, S.; Roy, S.; Banerjee, J.; Hussain, S.-R.A.; Khanna, S.; Meenakshisundaram, G.; Kuppusamy, P.; Friedman, A.; Sen, C.K. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. USA 2010, 107, 6976–6981. [Google Scholar] [CrossRef] [PubMed]
- Caporali, A.; Meloni, M.; Völlenkle, C.; Bonci, D.; Sala-Newby, G.B.; Addis, R.; Spinetti, G.; Losa, S.; Masson, R.; Baker, A.H. Deregulation of microRNA-503 contributes to diabetes mellitus–induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation 2011, 123, 282–291. [Google Scholar] [CrossRef]
- Spinetti, G.; Fortunato, O.; Caporali, A.; Shantikumar, S.; Marchetti, M.; Meloni, M.; Descamps, B.; Floris, I.; Sangalli, E.; Vono, R. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ. Res. 2013, 112, 335–346. [Google Scholar] [CrossRef]
- Spinetti, G.; Cordella, D.; Fortunato, O.; Sangalli, E.; Losa, S.; Gotti, A.; Carnelli, F.; Rosa, F.; Riboldi, S.; Sessa, F. Global remodeling of the vascular stem cell niche in bone marrow of diabetic patients: Implication of the microRNA-155/FOXO3a signaling pathway. Circ. Res. 2013, 112, 510–522. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Tan, C.; Yue, X.; Zhao, Y.; Peng, J.; Wang, X.; Laddha, S.V.; Chan, C.S.; Zheng, S. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget 2016, 7, 8783. [Google Scholar] [CrossRef]
- Fan, G.; Xu, P.; Tu, P. MiR-1827 functions as a tumor suppressor in lung adenocarcinoma by targeting MYC and FAM83F. J. Cell. Biochem. 2020, 121, 1675–1689. [Google Scholar] [CrossRef]
- San Ho, C.; Noor, S.M.; Nagoor, N.H. MiR-378 and MiR-1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J. Cancer 2018, 9, 331. [Google Scholar]

| Discovery Group (n = 23) | Confirmation Group (n = 52) | Validation Group (n = 20) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| non-PAD (n = 10) | CLTI (n = 13) | p-Value α | non-PAD (n = 20) | CLTI (n = 32) | p-Value α | non-PAD (n = 10) | CLTI (n = 10) | p-Value α | |
| Mean (SD) † | |||||||||
| Age | 62.4(6.2) | 70.3(8.6) | 0.02 | 55.7(15.3) | 71.0(7.7) | <0.001 | 77.9(5.6) | 78.6(6.4) | 0.733 |
| ABI | 1.0 (0.1) | 0.4 (0.1) | <0.001 | 1.1 (0.1) | 0.4 (0.1) | <0.001 | 1.1 (0.1) | 0.5 (0.1) | <0.001 |
| Frequency (%) ‡ | |||||||||
| Sex (male) | 5 (50) | 9 (69) | 0.417 | 8 (40) | 18 (58) | 0.258 | 9 (90) | 8 (80) | 1.00 |
| Hypertension | 5 (50) | 10 (77) | 0.221 | 3 (15) | 26 (84) | <0.001 | 8 (80) | 9 (90) | 1.00 |
| Hypercholesterolemia | 5 (50) | 12 (92) | 0.052 | 6 (30) | 26 (84) | <0.001 | 5 (50) | 9 (90) | 0.141 |
| Diabetes | 0 (0) | 1 (8) | 1.00 | 2 (10) | 14 (45) | 0.012 | 3 (30) | 2 (20) | 1.00 |
| Smoking History | 6 (60) | 12 (92) | 0.127 | 10 (50) | 25 (81) | 0.031 | 5 (50) | 8 (80) | 0.350 |
| Coronary artery disease | 0 (0) | 8 (62) | 0.003 | 1 (5) | 18 (58) | <0.001 | 3 (30) | 7 (70) | 0.179 |
| Stroke | 0 (0) | 0 (0) | NA | 1 (5) | 6 (19) | 0.229 | 0 (0) | 3 (30) | 0.211 |
| Medication (%) ‡ | |||||||||
| Statin | 4 (40) | 13 (100) | 0.002 | 6 (30) | 24 (77) | 0.005 | 5 (50) | 9 (90) | 0.141 |
| ACEi/Arb | 3 (30) | 8 (62) | 0.214 | 3 (15) | 20 (65) | 0.001 | 7 (70) | 8 (80) | 1.00 |
| Beta Blocker | 1 (10) | 4 (31) | 0.339 | 1 (5) | 11 (36) | 0.017 | 3 (30) | 7 (70) | 0.179 |
| Insulin | 0 (0) | 0 (0) | NA | 0 (0) | 4 (13) | 0.145 | 0 (0) | 2 (20) | 0.474 |
| GO Term | Overlapping Gene IDs | p-Value |
|---|---|---|
| Positive regulation of transport | CASP8; HCAR2; AHSG; HCLS1; DAB2; PPID; BCL2 | 4.34 × 10−5 |
| Regulation of protein localization | CASP8; HCAR2; HCLS1; DAB2; LCP1; PPID; BCL2 | 4.80 × 10−5 |
| Regulation of transport | CASP8; THBS1; HCAR2; AHSG; HCLS1; DAB2; LCP1; PPID; BCL2 | 5.44 × 10−5 |
| Response to hormone | BTG2; CASP8; THBS1; AHSG; HCLS1; DAB2; BCL2 | 5.50 × 10−5 |
| Negative regulation of metabolic process | TNFSF13; BTG2; THBS1; HCAR2; AHSG; HCLS1; DAB2; PPID; GPRC5A; CTDSP2; BCL2 | 6.89 × 10−5 |
| Pathway | Protein Members | Source | p-Value |
|---|---|---|---|
| Hydroxycarboxylic acid-binding receptors | HCAR3; HCAR2 | Reactome | 6.07 × 10−6 |
| P53 signaling pathway—Homo sapiens (human) | BCL2; CASP8; THBS1 | KEGG | 0.0001 |
| Nanomaterial induced apoptosis | CASP8; BCL2 | Wikipathways | 0.0003 |
| Ceramide signaling pathway | CASP8; BCL2 | BioCarta | 0.0010 |
| Apoptosis—multiple species—Homo sapiens (human) | CASP8; BCL2 | KEGG | 0.0010 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syed, M.H.; Zamzam, A.; Valencia, J.; Khan, H.; Jain, S.; Singh, K.K.; Abdin, R.; Qadura, M. MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia. Diagnostics 2020, 10, 230. https://doi.org/10.3390/diagnostics10040230
Syed MH, Zamzam A, Valencia J, Khan H, Jain S, Singh KK, Abdin R, Qadura M. MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia. Diagnostics. 2020; 10(4):230. https://doi.org/10.3390/diagnostics10040230
Chicago/Turabian StyleSyed, Muzammil H., Abdelrahman Zamzam, Jason Valencia, Hamzah Khan, Shubha Jain, Krishna K. Singh, Rawand Abdin, and Mohammad Qadura. 2020. "MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia" Diagnostics 10, no. 4: 230. https://doi.org/10.3390/diagnostics10040230
APA StyleSyed, M. H., Zamzam, A., Valencia, J., Khan, H., Jain, S., Singh, K. K., Abdin, R., & Qadura, M. (2020). MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia. Diagnostics, 10(4), 230. https://doi.org/10.3390/diagnostics10040230









