Predictive Analyses of Prognostic-Related Immune Genes and Immune Infiltrates for Glioblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Data Preparation
2.2. Screening of Prognostic-Related Immune Genes
2.3. Construction of Prognostic Immune Genes and Immune Gene Transcription Factor Regulatory Networks
2.4. Establishment of Immune Gene Model
2.5. Survival Analysis
2.6. Receiver Operating Characteristic (ROC) Curve
2.7. Risk Curve
2.8. Independent Prognostic Analysis
2.9. Clinical Correlation Analysis
2.10. Correlation Analysis of Immune Cell Infiltration and Risk Score
3. Results
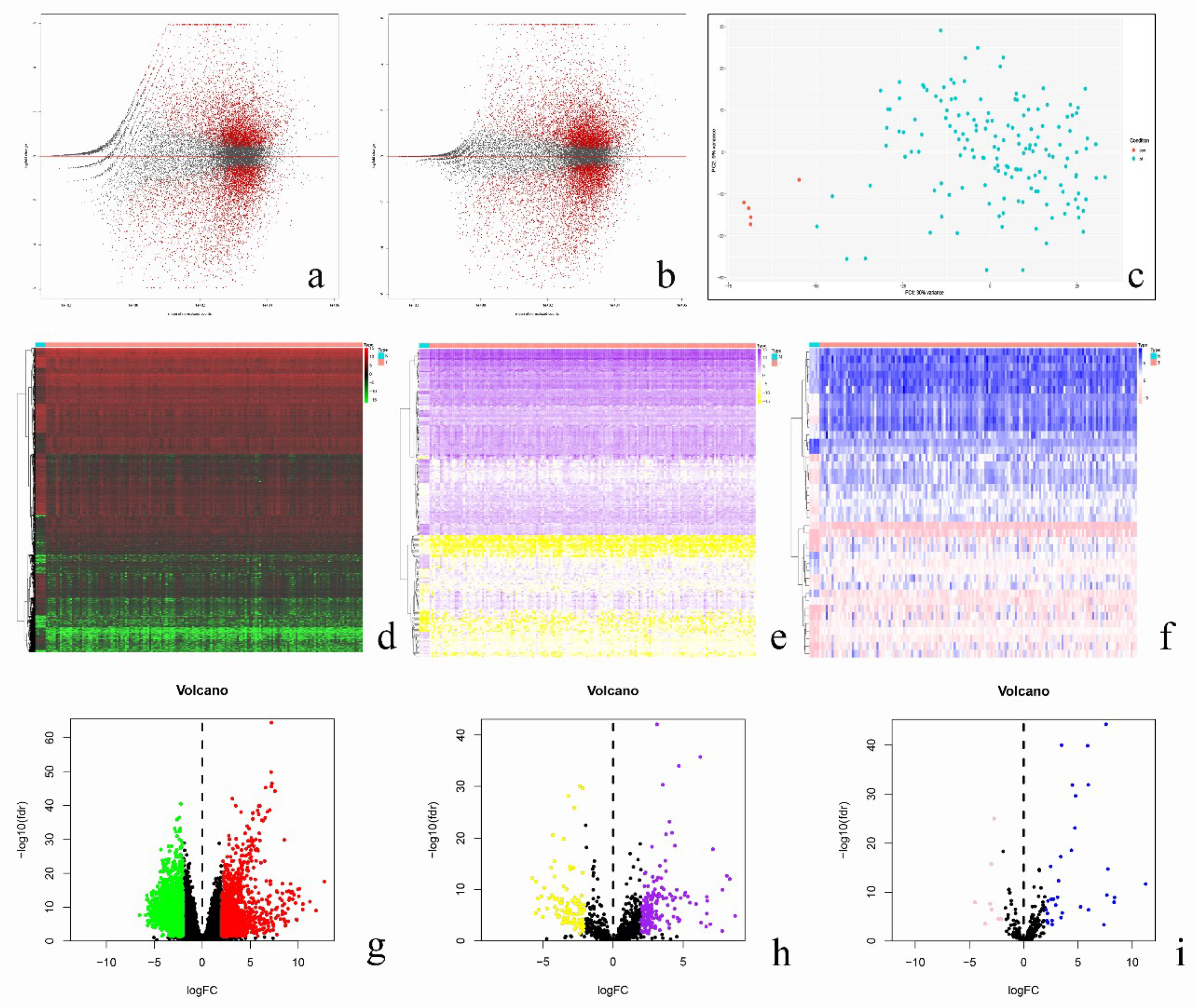
3.1. Identification of Prognostic-Related Immune Genes
3.2. Analysis of Regulatory Network of Immune Transcription Factors and Prognostic-Related Immune Genes
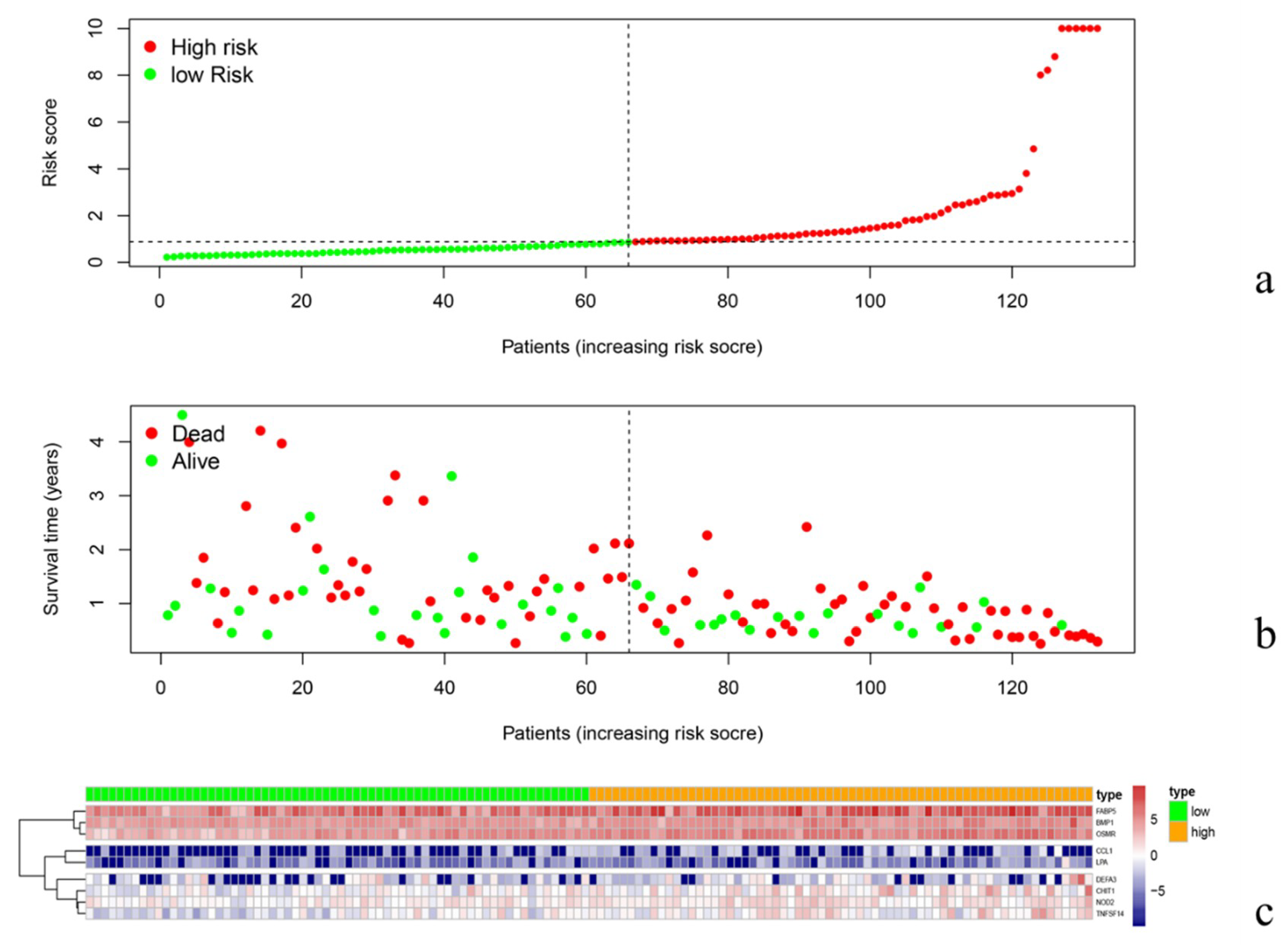
3.3. Validation of Prognosis Prediction Model and Survival Analysis
3.4. Risk Curve Analysis
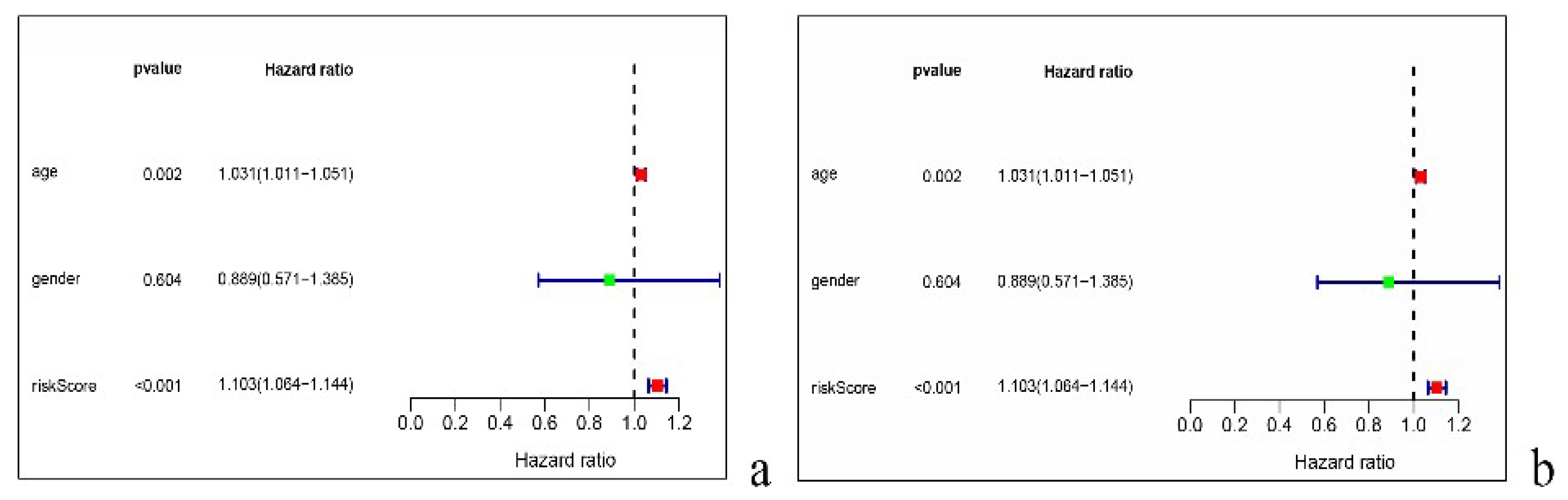
3.5. Univariate and Multivariate Independent Prognostic Analysis
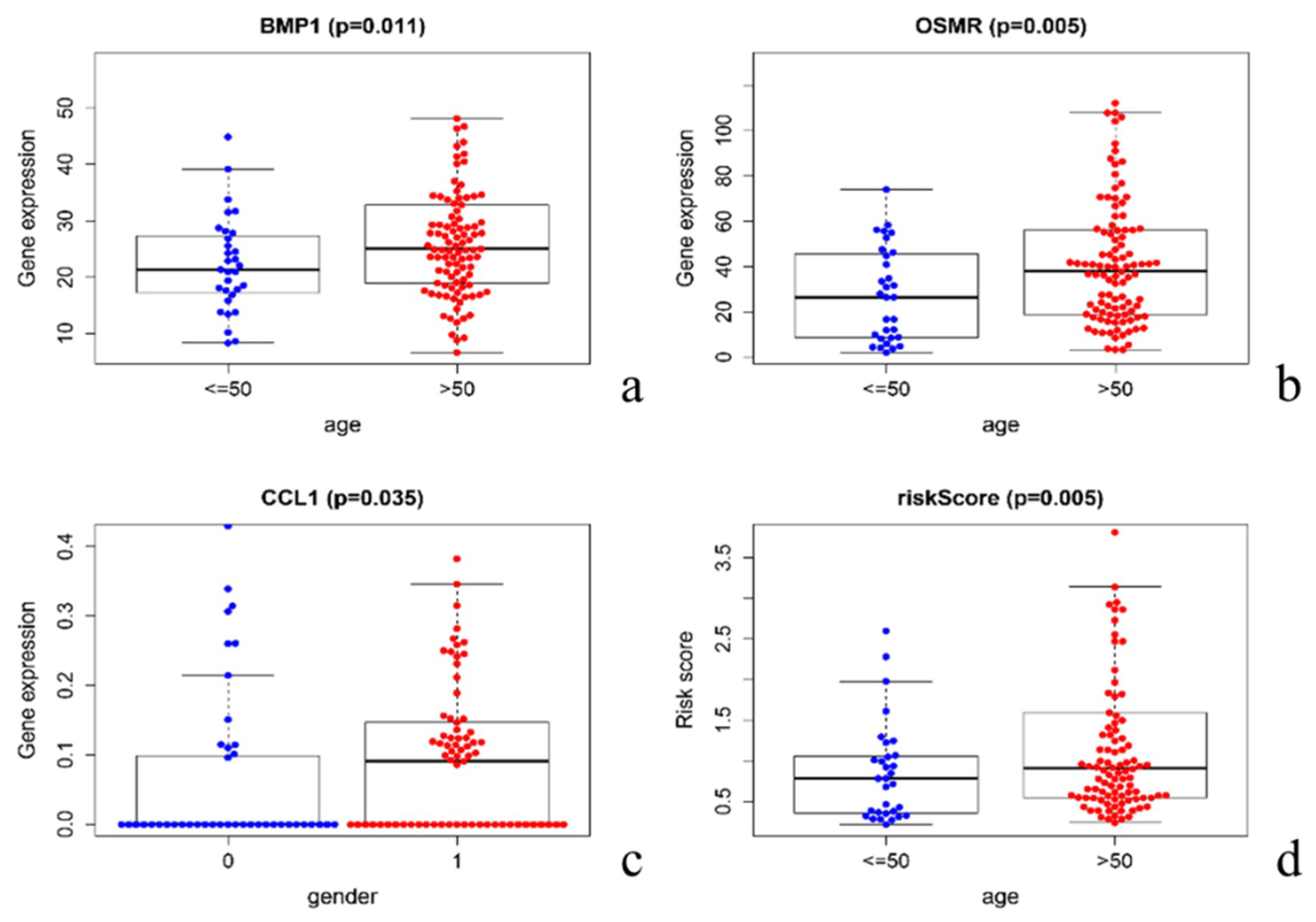
3.6. Clinical Relevance Verification
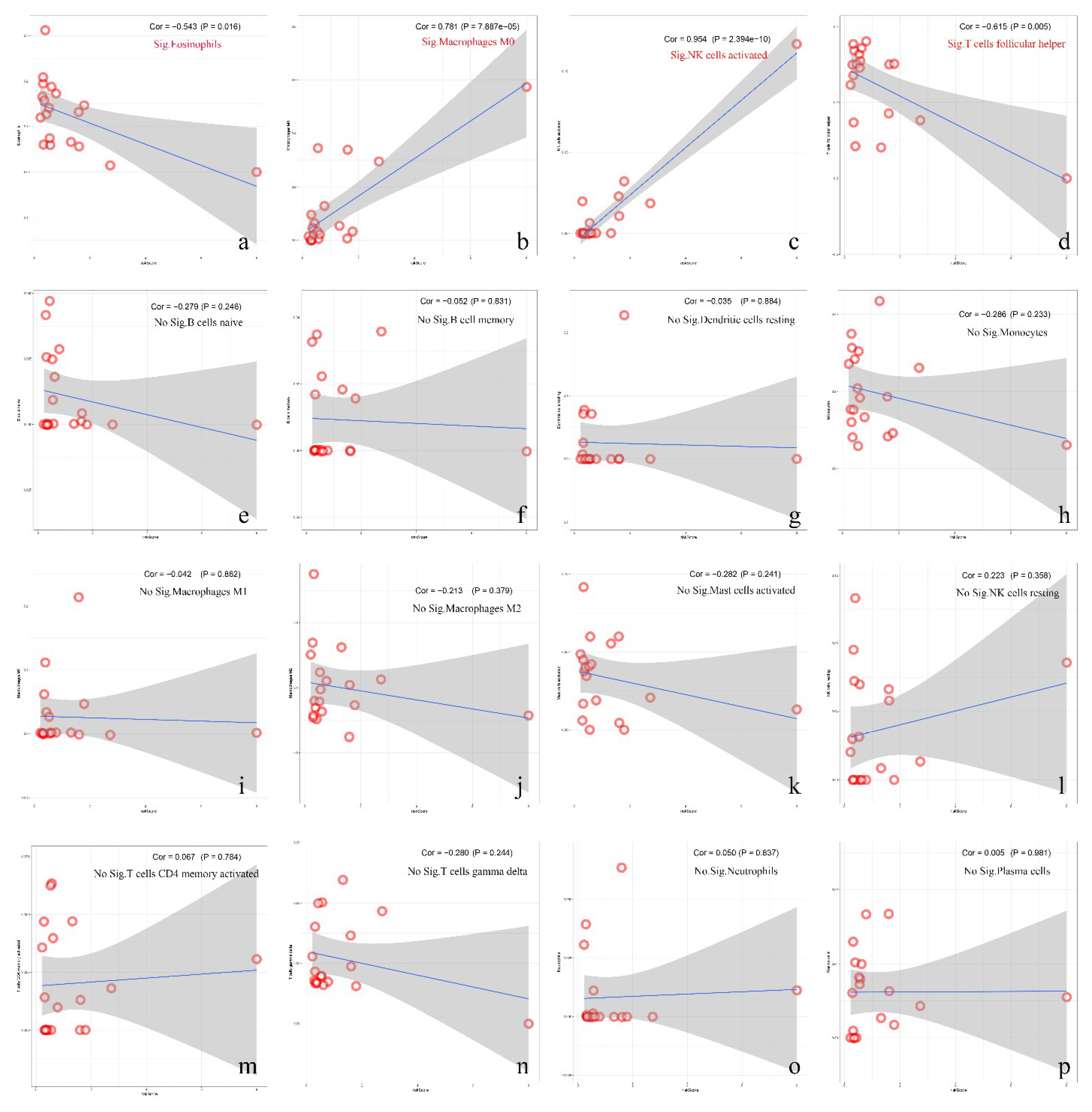
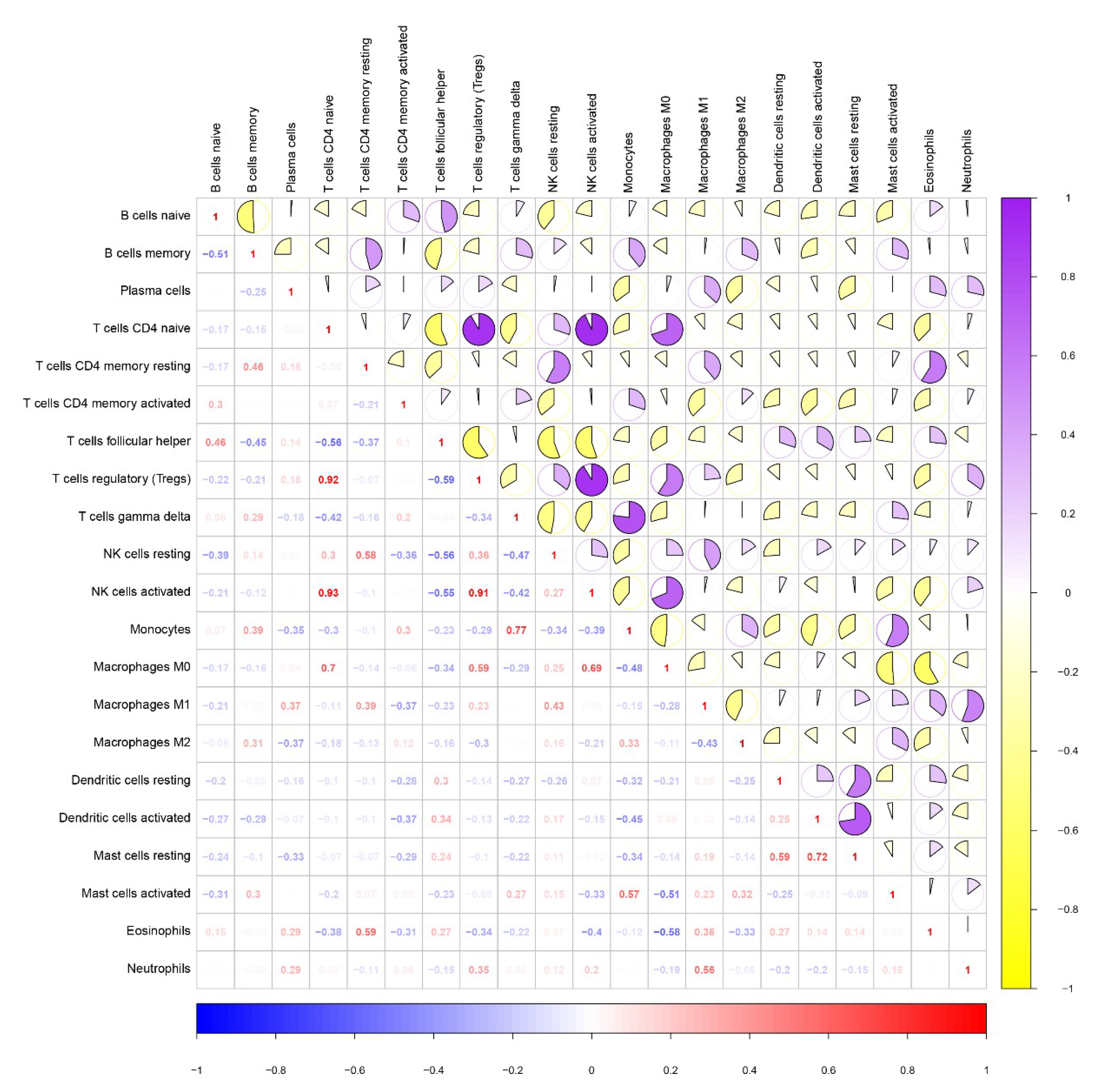
3.7. Correlation between Immune Cell Infiltration and Risk Score
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Perry, J.R.; Laperriere, N.; O’Callaghan, C.J.; Brandes, A.A.; Menten, J.; Phillips, C.; Fay, M.; Nishikawa, R.; Cairncross, J.G.; Roa, W.; et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N. Engl. J. Med. 2017, 376, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Verhaak, R.G.W.; Al-dape, K.D.; Yung, W.K.A.; Salama, S.R.; Cooper, L.A.D.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; Morozova, O.; et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; Decarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment (vol. 32, pg 42, 2017). Cancer Cell 2018, 33, 152. [Google Scholar] [CrossRef]
- Suva, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Senbabaoglu, Y.; Gejman, R.S.; Winer, A.G.; Liu, M.; Van Allen, E.M.; de Velasco, G.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A.; et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, D.A.; Chang, A.L.; Dey, M.; Balyasnikova, I.V.; Kim, C.K.; Tobias, A.; Cheng, Y.; Kim, J.W.; Qiao, J.; Zhang, L.; et al. Durable Therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer Res. 2014, 20, 5290–5301. [Google Scholar] [CrossRef]
- Heimberger, A.B.; Sun, W.; Hussain, S.F.; Dey, M.; Crutcher, L.; Aldape, K.; Gilbert, M.; Hassenbusch, S.J.; Sawaya, R.; Schmittling, B.; et al. Immunological responses in a patient with glioblastoma multiforme treated with sequential courses of temozolomide and immunotherapy: Case study. Neuro Oncol. 2008, 10, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.L.; Tussiwand, R.; Murphy, K.M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013, 13, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Barbus, S.; Tews, B.; Karra, D.; Hahn, M.; Radlwimmer, B.; Delhomme, N.; Hartmann, C.; Felsberg, J.; Krex, D.; Schackert, G.; et al. Differential retinoic acid signaling in tumors of long-and short-term glioblastoma survivors. JNCI J. Natl. Cancer Inst. 2011, 103, 598–601. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, D.; Cheng, S.; Fan, K.; Sheng, L.; Zhang, J.; Feng, B.; Xu, Z. The correlation of bone morphogenetic protein 2 with poor prognosis in glioma patients. Tumor Biol. 2014, 35, 11091–11095. [Google Scholar] [CrossRef]
- Jahani-Asl, A.; Yin, H.; Soleimani, V.D.; Haque, T.; Luchman, H.A.; Chang, N.C.; Sincennes, M.-C.; Puram, S.V.; Scott, A.M.; Lorimer, I.A.J.; et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat. Neurosci. 2016, 19, 798–806. [Google Scholar] [CrossRef]
- Mansfield, A.S.; Nevala, W.K.; Dronca, R.S.; Leontovich, A.A.; Shuster, L.; Markovic, S.N. Normal ageing is associated with an increase in Th2 cells, MCP-1 (CCL1) and RANTES (CCL5), with differences in sCD40L and PDGF-AA between sexes. Clin. Exp. Immunol. 2012, 170, 186–193. [Google Scholar] [CrossRef]
- Tepper, R.I.; Pattengale, P.K.; Leder, P. Murine interleukin-4 displays potent anti-tumor activity invivo. Cell 1989, 57, 503–512. [Google Scholar] [CrossRef]
- Hayes, R.L.; Arbit, E.; Odaimi, M.; Pannullo, S.; Scheff, R.; Kravchinskiy, D.; Zaroulis, C. Adoptive cellular immunotherapy for the treatment of malignant gliomas. Crit. Rev. Oncol. Hematol. 2001, 39, 31–42. [Google Scholar] [CrossRef]
- Graeber, M.B.; Scheithauer, B.W.; Kreutzberg, G.W. Microglia in brain tumors. Glia 2002, 40, 252–259. [Google Scholar] [CrossRef]
- Gutmann, D.H.; Kettenmann, H. Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron 2019, 104, 442–449. [Google Scholar] [CrossRef]
- Prosniak, M.; Harshyne, L.A.; Andrews, D.W.; Kenyon, L.C.; Bedelbaeva, K.; Apanasovich, T.V.; Heber-Katz, E.; Curtis, M.T.; Cotzia, P.; Hooper, D.C. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin. Cancer Res. 2013, 19, 3776–3786. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain tumor microenvironment and host state: Implications for immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef] [PubMed]
- Wischhusen, J.; Friese, M.A.; Mittelbronn, M.; Meyermann, R.; Weller, M. HLA-E protects glioma cells from NKG2D-mediated immune responses in vitro: Implications for immune escape in vivo. J. Neuropathol. Exp. Neurol. 2005, 64, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Wiendl, H.; Mitsdoerffer, M.; Hofmeister, V.; Wischhusen, J.; Bornemann, A.; Meyermann, R.; Weiss, E.H.; Melms, A.; Weller, M. A functional role of HLA-G expression in human gliomas: An alternative strategy of immune escape. J. Immunol. 2002, 168, 4772–4780. [Google Scholar] [CrossRef]
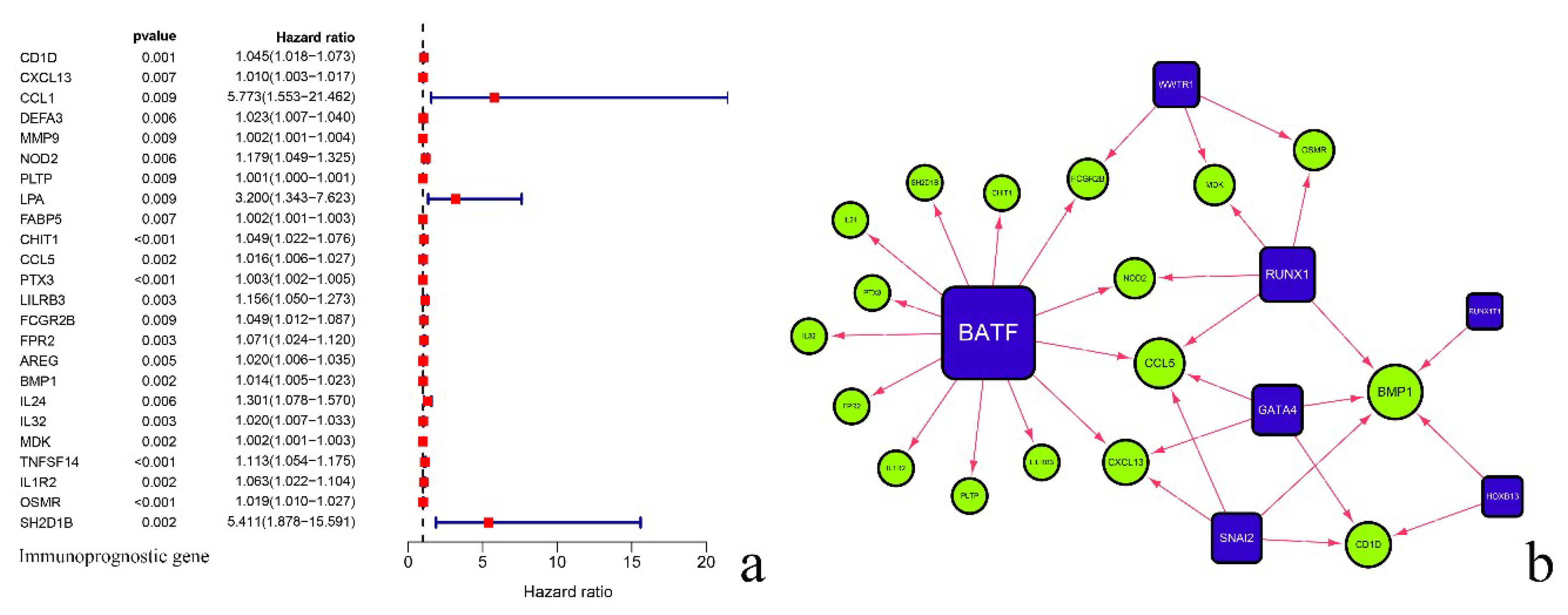


| Transcription Factor | ImmuneGene | Correlation | p Value | Regulation | Transcription Factor | Immunegene | Correlation | p Value | Regulation |
|---|---|---|---|---|---|---|---|---|---|
| SNAI2 | CD1D | 0.70 | 3.15 × 10−21 | positive | BATF | CXCL13 | 0.43 | 2.09 × 10−7 | positive |
| CXCL13 | 0.54 | 1.72 × 10−11 | positive | NOD2 | 0.77 | 1.04 × 10−27 | positive | ||
| CCL5 | 0.63 | 3.57 × 10−16 | positive | PLTP | 0.73 | 1.20 × 10−23 | positive | ||
| BMP1 | 0.80 | 9.53 × 10−31 | positive | CHIT1 | 0.53 | 5.82 × 10−11 | positive | ||
| GATA4 | CD1D | 0.64 | 6.70 × 10−17 | positive | CCL5 | 0.54 | 9.59 × 10−12 | positive | |
| CXCL13 | 0.45 | 3.99 × 10−8 | positive | PTX3 | 0.62 | 1.67 × 10−15 | positive | ||
| CCL5 | 0.46 | 1.46 × 10−8 | positive | LILRB3 | 0.72 | 1.43 × 10−22 | positive | ||
| BMP1 | 0.64 | 6.94 × 10−17 | positive | FCGR2B | 0.60 | 1.56 × 10−14 | positive | ||
| HOXB13 | CD1D | 0.55 | 3.04 × 10−12 | positive | FPR2 | 0.71 | 2.94 × 10−22 | positive | |
| BMP1 | 0.53 | 2.37 × 10−11 | positive | IL24 | 0.48 | 4.35 × 10−9 | positive | ||
| RUNX1 | NOD2 | 0.48 | 3.11 × 10−9 | positive | IL32 | 0.40 | 1.73 × 10−6 | positive | |
| CCL5 | 0.49 | 1.33 × 10−9 | positive | IL1R2 | 0.45 | 3.88 × 10−8 | positive | ||
| BMP1 | 0.55 | 6.08 × 10−12 | positive | SH2D1B | 0.45 | 2.91 × 10−8 | positive | ||
| MDK | 0.49 | 1.74 × 10−9 | positive | WWTR1 | FCGR2B | 0.40 | 1.20 × 10−6 | positive | |
| OSMR | 0.48 | 5.14 × 10−9 | positive | MDK | 0.40 | 1.40 × 10−6 | positive | ||
| RUNX1T1 | BMP1 | 0.42 | 4.02 × 10−7 | positive | OSMR | 0.44 | 1.13 × 10−7 | positive |
| ImmuneGene | coef | HR | HR.95L | HR.95H | p Value |
|---|---|---|---|---|---|
| CCL1 | 2.40 | 11.09 | 2.93 | 41.90 | 0.00 |
| DEFA3 | 0.03 | 1.03 | 1.01 | 1.05 | 0.00 |
| NOD2 | −0.15 | 0.86 | 0.72 | 1.02 | 0.09 |
| LPA | 1.50 | 4.52 | 1.80 | 11.35 | 0.00 |
| FABP5 | 0.00 | 1.00 | 0.99 | 1.00 | 0.05 |
| CHIT1 | 0.06 | 1.07 | 1.03 | 1.10 | 0.00 |
| BMP1 | 0.01 | 1.01 | 1.00 | 1.02 | 0.00 |
| TNFSF14 | 0.08 | 1.08 | 1.00 | 1.17 | 0.03 |
| OSMR | 0.02 | 1.01 | 1.00 | 1.03 | 0.00 |
| Risk = High | Risk = Low | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (year) | n.Risk | n.Event | Survival (%) | Std.err | Lower 95% CI | Upper 95% CI | Time (year) | n.risk | n.event | Survival (%) | Std.err | Lower 95% CI | Upper 95% CI |
| 0.26 | 66.00 | 1.00 | 0.98 | 0.02 | 0.96 | 1.00 | 0.27 | 66.00 | 1.00 | 0.98 | 0.02 | 0.96 | 1.00 |
| 0.27 | 65.00 | 1.00 | 0.97 | 0.02 | 0.93 | 1.00 | 0.27 | 65.00 | 1.00 | 0.97 | 0.02 | 0.93 | 1.00 |
| 0.30 | 64.00 | 1.00 | 0.95 | 0.03 | 0.91 | 1.00 | 0.33 | 64.00 | 1.00 | 0.95 | 0.03 | 0.91 | 1.00 |
| 0.30 | 63.00 | 1.00 | 0.94 | 0.03 | 0.88 | 1.00 | 0.41 | 61.00 | 1.00 | 0.94 | 0.03 | 0.88 | 1.00 |
| 0.31 | 62.00 | 1.00 | 0.92 | 0.03 | 0.86 | 0.99 | 0.64 | 55.00 | 1.00 | 0.92 | 0.03 | 0.86 | 0.99 |
| 0.34 | 61.00 | 1.00 | 0.91 | 0.04 | 0.84 | 0.98 | 0.70 | 54.00 | 1.00 | 0.90 | 0.04 | 0.83 | 0.98 |
| 0.36 | 60.00 | 1.00 | 0.89 | 0.04 | 0.82 | 0.97 | 0.74 | 52.00 | 1.00 | 0.89 | 0.04 | 0.81 | 0.97 |
| 0.38 | 59.00 | 2.00 | 0.86 | 0.04 | 0.78 | 0.95 | 0.76 | 50.00 | 1.00 | 0.87 | 0.04 | 0.79 | 0.96 |
| 0.39 | 57.00 | 1.00 | 0.85 | 0.04 | 0.77 | 0.94 | 1.04 | 42.00 | 1.00 | 0.85 | 0.05 | 0.76 | 0.95 |
| 0.40 | 56.00 | 1.00 | 0.83 | 0.05 | 0.75 | 0.93 | 1.08 | 41.00 | 1.00 | 0.83 | 0.05 | 0.74 | 0.93 |
| 0.41 | 55.00 | 1.00 | 0.82 | 0.05 | 0.73 | 0.92 | 1.11 | 40.00 | 1.00 | 0.81 | 0.05 | 0.71 | 0.92 |
| 0.42 | 54.00 | 1.00 | 0.80 | 0.05 | 0.71 | 0.91 | 1.11 | 39.00 | 1.00 | 0.79 | 0.06 | 0.69 | 0.90 |
| 0.43 | 53.00 | 1.00 | 0.79 | 0.05 | 0.70 | 0.89 | 1.15 | 38.00 | 2.00 | 0.75 | 0.06 | 0.64 | 0.87 |
| 0.45 | 52.00 | 1.00 | 0.77 | 0.05 | 0.68 | 0.88 | 1.21 | 36.00 | 1.00 | 0.72 | 0.06 | 0.61 | 0.86 |
| 0.48 | 49.00 | 2.00 | 0.74 | 0.05 | 0.64 | 0.86 | 1.23 | 34.00 | 2.00 | 0.68 | 0.06 | 0.57 | 0.82 |
| 0.49 | 47.00 | 1.00 | 0.73 | 0.06 | 0.62 | 0.84 | 1.24 | 31.00 | 2.00 | 0.64 | 0.07 | 0.52 | 0.79 |
| 0.61 | 38.00 | 1.00 | 0.71 | 0.06 | 0.60 | 0.83 | 1.32 | 27.00 | 1.00 | 0.61 | 0.07 | 0.49 | 0.77 |
| 0.62 | 37.00 | 1.00 | 0.69 | 0.06 | 0.58 | 0.81 | 1.33 | 26.00 | 1.00 | 0.59 | 0.07 | 0.47 | 0.75 |
| 0.63 | 36.00 | 1.00 | 0.67 | 0.06 | 0.56 | 0.80 | 1.34 | 25.00 | 1.00 | 0.57 | 0.07 | 0.44 | 0.73 |
| 0.66 | 35.00 | 1.00 | 0.65 | 0.06 | 0.54 | 0.78 | 1.38 | 24.00 | 1.00 | 0.54 | 0.07 | 0.42 | 0.71 |
| 0.74 | 33.00 | 1.00 | 0.63 | 0.06 | 0.52 | 0.76 | 1.46 | 23.00 | 1.00 | 0.52 | 0.07 | 0.39 | 0.69 |
| 0.82 | 27.00 | 1.00 | 0.61 | 0.06 | 0.49 | 0.75 | 1.47 | 22.00 | 1.00 | 0.50 | 0.07 | 0.37 | 0.66 |
| 0.86 | 26.00 | 1.00 | 0.58 | 0.07 | 0.47 | 0.73 | 1.49 | 21.00 | 1.00 | 0.47 | 0.07 | 0.35 | 0.64 |
| 0.87 | 25.00 | 1.00 | 0.56 | 0.07 | 0.44 | 0.71 | 1.64 | 19.00 | 1.00 | 0.45 | 0.07 | 0.32 | 0.62 |
| 0.89 | 24.00 | 1.00 | 0.54 | 0.07 | 0.42 | 0.69 | 1.78 | 18.00 | 1.00 | 0.42 | 0.07 | 0.30 | 0.60 |
| 0.90 | 23.00 | 1.00 | 0.51 | 0.07 | 0.39 | 0.67 | 1.85 | 17.00 | 1.00 | 0.40 | 0.07 | 0.28 | 0.57 |
| 0.91 | 22.00 | 1.00 | 0.49 | 0.07 | 0.37 | 0.65 | 2.02 | 15.00 | 2.00 | 0.34 | 0.07 | 0.23 | 0.52 |
| 0.92 | 21.00 | 1.00 | 0.47 | 0.07 | 0.35 | 0.63 | 2.11 | 13.00 | 1.00 | 0.32 | 0.07 | 0.20 | 0.50 |
| 0.94 | 20.00 | 1.00 | 0.44 | 0.07 | 0.32 | 0.61 | 2.12 | 12.00 | 1.00 | 0.29 | 0.07 | 0.18 | 0.47 |
| 0.94 | 19.00 | 1.00 | 0.42 | 0.07 | 0.30 | 0.58 | 2.41 | 11.00 | 1.00 | 0.27 | 0.07 | 0.16 | 0.44 |
| 0.98 | 18.00 | 1.00 | 0.40 | 0.07 | 0.28 | 0.56 | 2.81 | 9.00 | 1.00 | 0.24 | 0.07 | 0.13 | 0.41 |
| 0.99 | 17.00 | 2.00 | 0.35 | 0.07 | 0.24 | 0.52 | 2.91 | 8.00 | 2.00 | 0.18 | 0.06 | 0.09 | 0.35 |
| 0.99 | 15.00 | 1.00 | 0.33 | 0.07 | 0.22 | 0.49 | 3.38 | 5.00 | 1.00 | 0.14 | 0.06 | 0.06 | 0.32 |
| 1.06 | 13.00 | 1.00 | 0.30 | 0.07 | 0.19 | 0.47 | 3.97 | 4.00 | 1.00 | 0.11 | 0.05 | 0.04 | 0.29 |
| 1.08 | 12.00 | 1.00 | 0.28 | 0.07 | 0.17 | 0.44 | 4.00 | 3.00 | 1.00 | 0.07 | 0.05 | 0.02 | 0.25 |
| 1.13 | 11.00 | 1.00 | 0.25 | 0.07 | 0.15 | 0.42 | 4.21 | 2.00 | 1.00 | 0.04 | 0.03 | 0.01 | 0.25 |
| 1.17 | 9.00 | 1.00 | 0.22 | 0.06 | 0.13 | 0.39 | |||||||
| 1.28 | 8.00 | 1.00 | 0.20 | 0.06 | 0.11 | 0.36 | |||||||
| 1.33 | 6.00 | 1.00 | 0.16 | 0.06 | 0.08 | 0.33 | |||||||
| 1.50 | 4.00 | 1.00 | 0.12 | 0.06 | 0.05 | 0.30 | |||||||
| 1.58 | 3.00 | 1.00 | 0.08 | 0.05 | 0.02 | 0.27 | |||||||
| 2.27 | 2.00 | 1.00 | 0.04 | 0.04 | 0.01 | ||||||||
| 2.42 | 1.00 | 1.00 | 0.00 | ||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, P.; Chai, Y.; Zhao, H.; Wang, G. Predictive Analyses of Prognostic-Related Immune Genes and Immune Infiltrates for Glioblastoma. Diagnostics 2020, 10, 177. https://doi.org/10.3390/diagnostics10030177
Liang P, Chai Y, Zhao H, Wang G. Predictive Analyses of Prognostic-Related Immune Genes and Immune Infiltrates for Glioblastoma. Diagnostics. 2020; 10(3):177. https://doi.org/10.3390/diagnostics10030177
Chicago/Turabian StyleLiang, Ping, Yi Chai, He Zhao, and Guihuai Wang. 2020. "Predictive Analyses of Prognostic-Related Immune Genes and Immune Infiltrates for Glioblastoma" Diagnostics 10, no. 3: 177. https://doi.org/10.3390/diagnostics10030177
APA StyleLiang, P., Chai, Y., Zhao, H., & Wang, G. (2020). Predictive Analyses of Prognostic-Related Immune Genes and Immune Infiltrates for Glioblastoma. Diagnostics, 10(3), 177. https://doi.org/10.3390/diagnostics10030177




