Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2 V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients’ Samples
2.2. Reference Samples
2.3. DNA Extraction
2.4. qPCR
2.5. ddPCR
2.6. Statistical Analysis
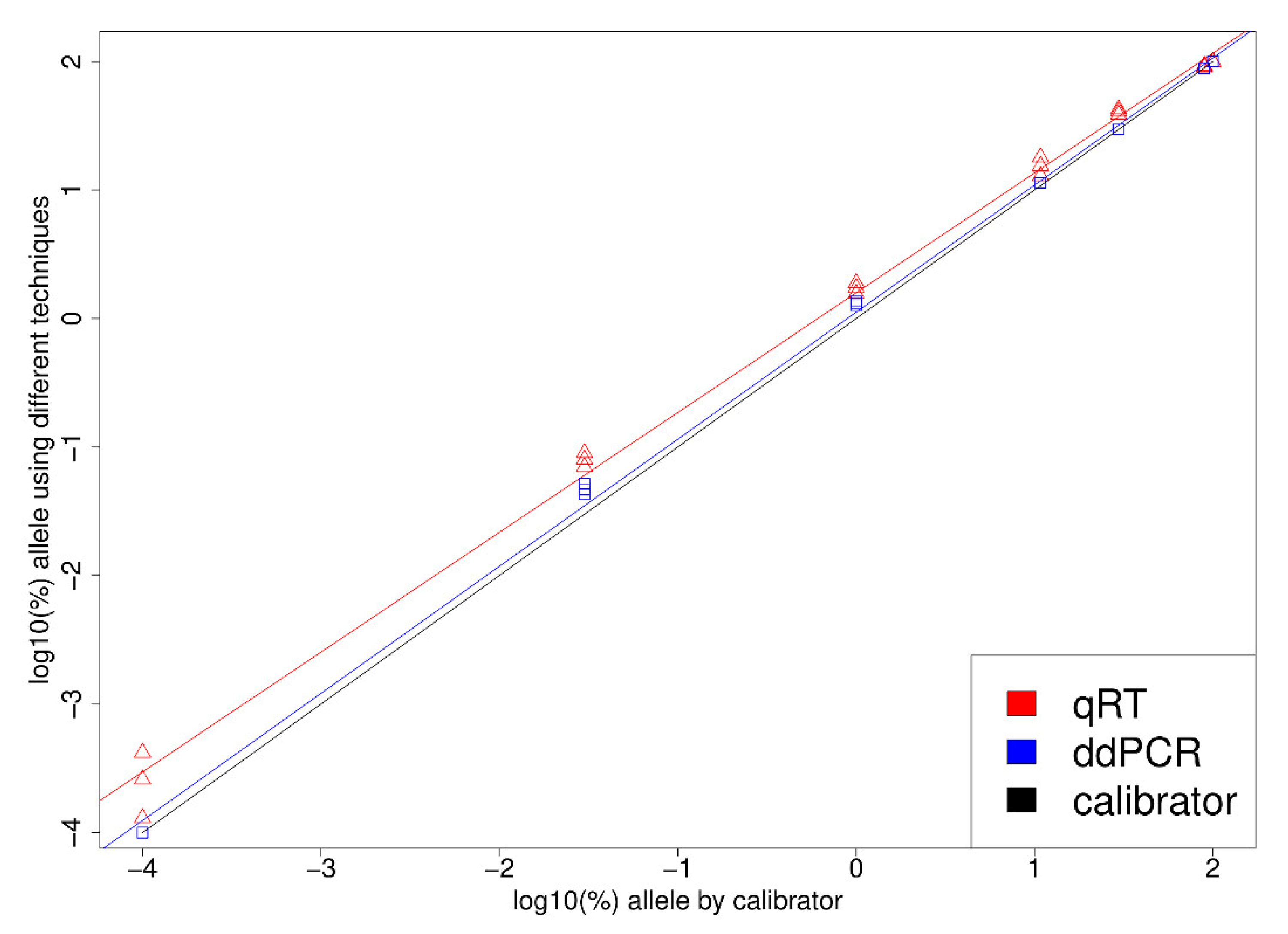
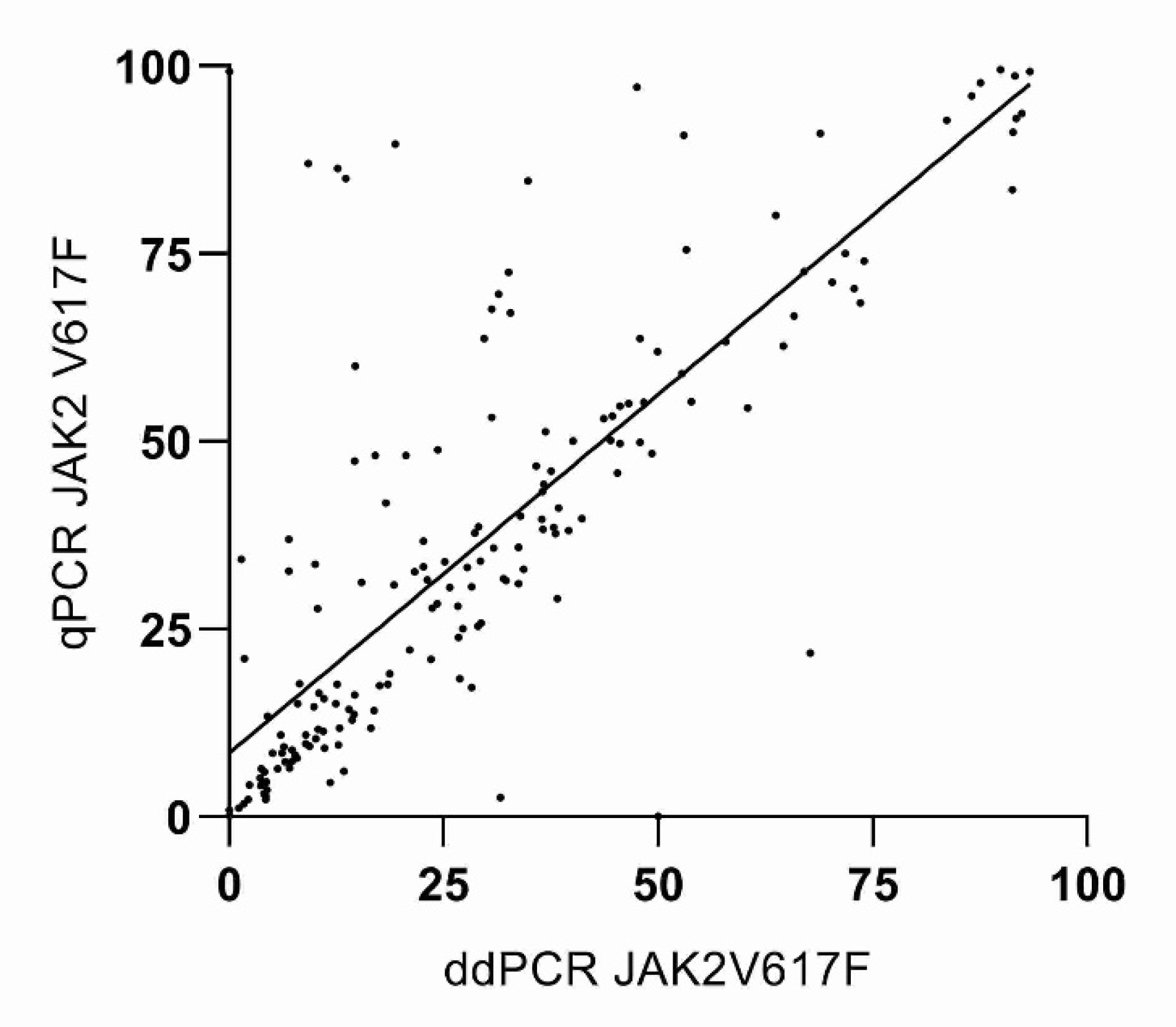
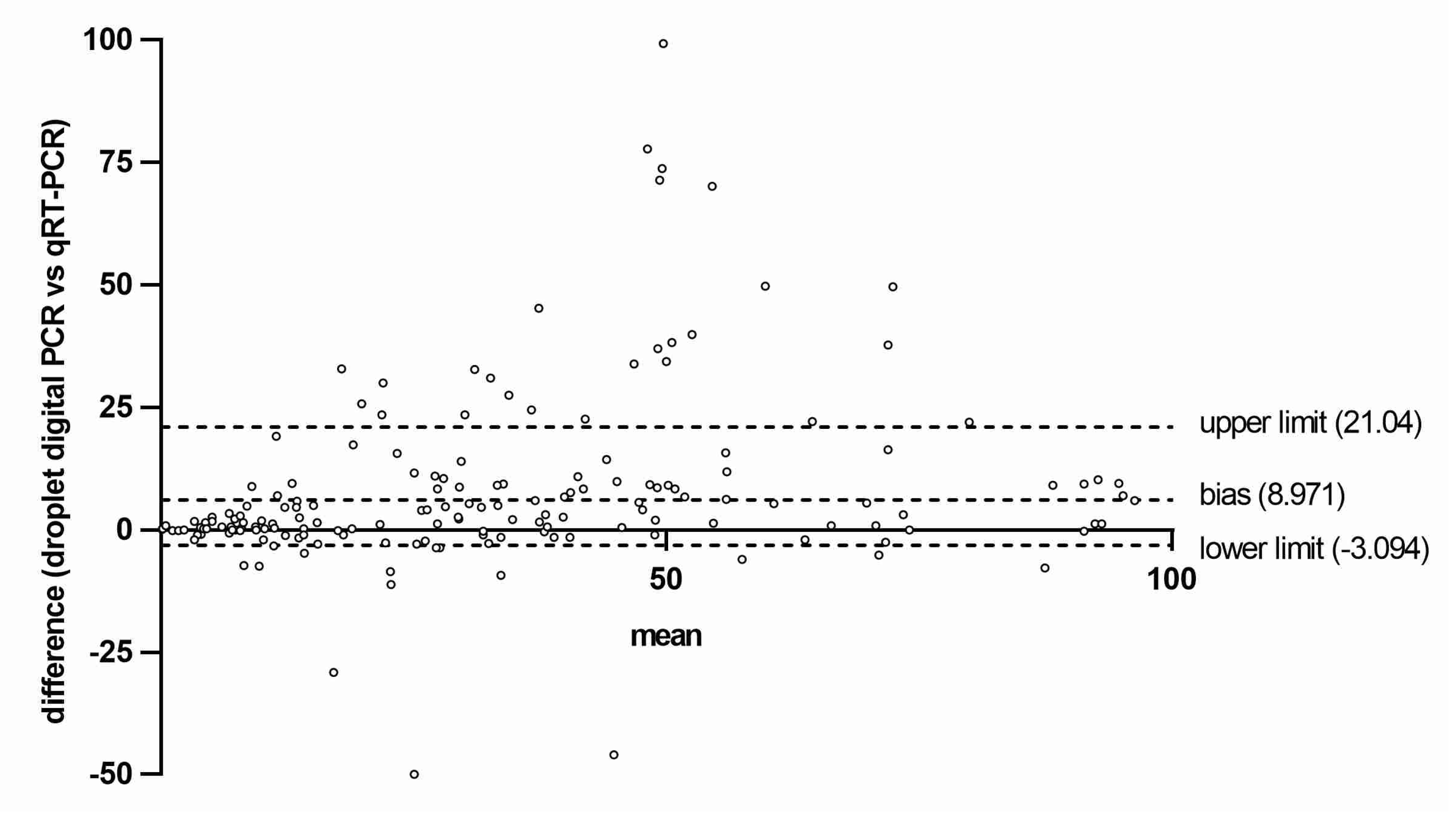
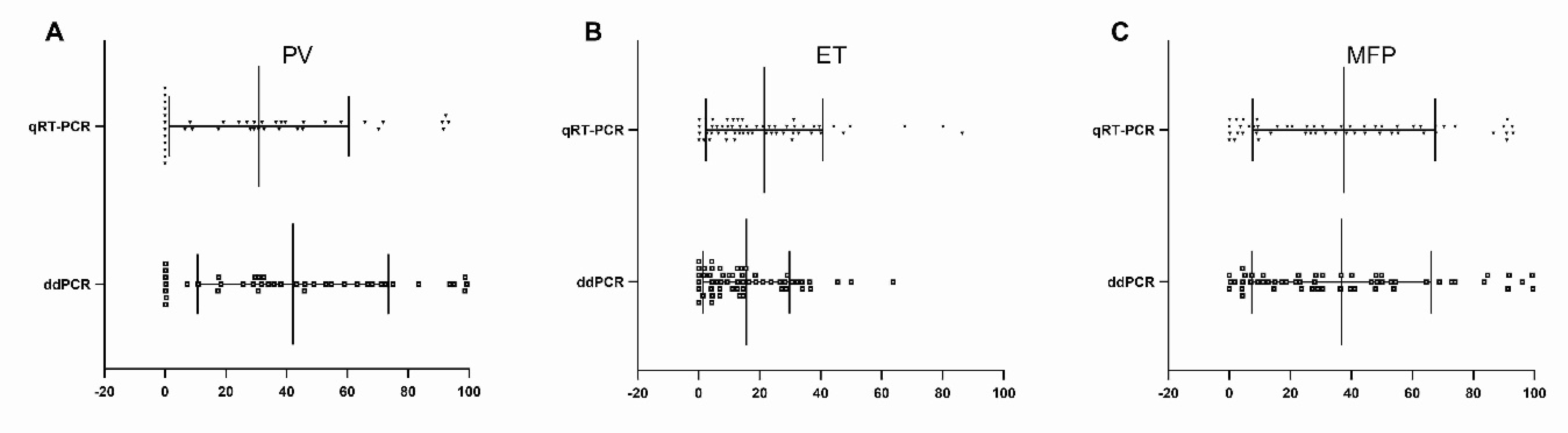
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Cancer Genome Project Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Verstovsek, S.; Silver, R.T.; Cross, N.C.P.; Tefferi, A. JAK2V617F mutational frequency in polycythemia vera: 100%, >90%, less? Leukemia 2006, 20, 2067. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Choi, H.-J.; Kim, Y.-K.; Kim, H.-J.; Shin, J.-H.; Suh, S.-P.; Ryang, D.-W.; Shin, M.-G. Allelic expression imbalance of JAK2 V617F mutation in BCR-ABL negative myeloproliferative neoplasms. PLoS ONE 2013, 8, e52518. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, F.; Rumi, E. Clinical relevance of JAK2 (V617F) mutant allele burden. Haematologica 2009, 94, 7–10. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Huang, J.; Finke, C.; Mesa, R.A.; Li, C.Y.; Wu, W.; Hanson, C.A.; Pardanani, A. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia 2008, 22, 756–761. [Google Scholar] [CrossRef]
- Langabeer, S.E.; Andrikovics, H.; Asp, J.; Bellosillo, B.; Carillo, S.; Haslam, K.; Kjaer, L.; Lippert, E.; Mansier, O.; Oppliger Leibundgut, E.; et al. MPN&MPNr-EuroNet Molecular diagnostics of myeloproliferative neoplasms. Eur. J. Haematol. 2015, 95, 270–279. [Google Scholar] [CrossRef]
- Campbell, P.J.; Griesshammer, M.; Döhner, K.; Döhner, H.; Kusec, R.; Hasselbalch, H.C.; Larsen, T.S.; Pallisgaard, N.; Giraudier, S.; Le Bousse-Kerdilès, M.-C.; et al. V617F mutation in JAK2 is associated with poorer survival in idiopathic myelofibrosis. Blood 2006, 107, 2098–2100. [Google Scholar] [CrossRef]
- Koren-Michowitz, M.; Landman, J.; Cohen, Y.; Rahimi-Levene, N.; Salomon, O.; Michael, M.; Amariglio, N.; Nagler, A. JAK2V617F allele burden is associated with transformation to myelofibrosis. Leuk. Lymphoma 2012, 53, 2210–2213. [Google Scholar] [CrossRef]
- Vannucchi, A.M.; Pieri, L.; Guglielmelli, P. JAK2 Allele Burden in the Myeloproliferative Neoplasms: Effects on Phenotype, Prognosis and Change with Treatment. Ther. Adv. Hematol. 2011, 2, 21–32. [Google Scholar] [CrossRef]
- Hayden, R.T.; Gu, Z.; Ingersoll, J.; Abdul-Ali, D.; Shi, L.; Pounds, S.; Caliendo, A.M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013, 51, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; Impera, L.; Minervini, C.F.; Brunetti, C.; Ricco, A.; et al. Droplet digital PCR assay for quantifying of CALR mutant allelic burden in myeloproliferative neoplasms. Ann. Hematol. 2016, 95, 1559–1560. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, M.; Follo, M.; Pfeifer, D.; von Bubnoff, N.; Duyster, J.; Bertz, H.; Finke, J. Sensitive and accurate quantification of JAK2 V617F mutation in chronic myeloproliferative neoplasms by droplet digital PCR. Ann. Hematol. 2016, 95, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Huggett, J.F.; Bushell, C.A.; Cowen, S.; Scott, D.J.; Foy, C.A. Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 2011, 83, 6474–6484. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Measurement in Medicine: The Analysis of Method Comparison Studies. Statisticians 1983, 32, 307. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar]
- Barosi, G.; Birgegard, G.; Finazzi, G.; Griesshammer, M.; Harrison, C.; Hasselbalch, H.C.; Kiladjian, J.-J.; Lengfelder, E.; McMullin, M.F.; Passamonti, F.; et al. Response criteria for essential thrombocythemia and polycythemia vera: Result of a European LeukemiaNet consensus conference. Blood 2009, 113, 4829–4833. [Google Scholar] [CrossRef]
- Cordua, S.; Kjaer, L.; Skov, V.; Pallisgaard, N.; Hasselbalch, H.C.; Ellervik, C. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood 2019, 134, 469–479. [Google Scholar] [CrossRef]
- Minnucci, G.; Amicarelli, G.; Salmoiraghi, S.; Spinelli, O.; Guinea Montalvo, M.L.; Giussani, U.; Adlerstein, D.; Rambaldi, A. A novel, highly sensitive and rapid allele-specific loop-mediated amplification assay for the detection of the JAK2V617F mutation in chronic myeloproliferative neoplasms. Haematologica 2012, 97, 1394–1400. [Google Scholar] [CrossRef][Green Version]
- Roma, C.; Esposito, C.; Rachiglio, A.M.; Pasquale, R.; Iannaccone, A.; Chicchinelli, N.; Franco, R.; Mancini, R.; Pisconti, S.; De Luca, A.; et al. Detection of EGFR mutations by TaqMan mutation detection assays powered by competitive allele-specific TaqMan PCR technology. Biomed. Res. Int. 2013, 2013, 385087. [Google Scholar] [CrossRef]
- McEvoy, A.C.; Wood, B.A.; Ardakani, N.M.; Pereira, M.; Pearce, R.; Cowell, L.; Robinson, C.; Grieu-Iacopetta, F.; Spicer, A.J.; Amanuel, B.; et al. Droplet digital PCR for mutation detection in formalin-fixed, paraffinembedded melanoma tissues: A comparison with Sanger sequencing and pyrosequencing. J. Mol. Diagn. 2018, 20, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, K.; Jing, C.; Cao, H.; Ma, R.; Wu, J. Comparison of droplet digital PCR and direct Sanger sequencing for the detection of the BRAFV600E mutation in papillary thyroid carcinoma. J. Clin. Lab. Anal. 2019, 33, e22902. [Google Scholar] [CrossRef] [PubMed]
- Nussenzveig, R.H.; Swierczek, S.I.; Jelinek, J.; Gaikwad, A.; Liu, E.; Verstovsek, S.; Prchal, J.F.; Prchal, J.T. Polycythemia vera is not initiated by JAK2V617F mutation. Exp. Hematol. 2007, 35, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Link-Lenczowska, D.; Pallisgaard, N.; Cordua, S.; Zawada, M.; Czekalska, S.; Krochmalczyk, D.; Kanduła, Z.; Sacha, T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 2018, 97, 2299–2308. [Google Scholar] [CrossRef]
- Fontanelli, G.; Baratè, C.; Ciabatti, E.; Guerrini, F.; Grassi, S.; Del Re, M.; Morganti, R.; Petrini, I.; Arici, R.; Barsotti, S.; et al. Real-Time PCR and Droplet Digital PCR: Two techniques for detection of the JAK2(V617F) mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int. J. Lab. Hematol. 2015, 37, 766–773. [Google Scholar] [CrossRef]
- Musto, P.; Simeon, V.; Guariglia, R.; Bianchino, G.; Grieco, V.; Nozza, F.; La Rocca, F.; Marziano, G.; Lalinga, A.V.; Fabiani, E.; et al. Myelodysplastic disorders carrying both isolated del(5q) and JAK2(V617F) mutation: Concise review, with focus on lenalidomide therapy. Oncol. Targets Ther. 2014, 7, 1043–1050. [Google Scholar] [CrossRef][Green Version]
- Lange, T.; Edelmann, A.; Siebolts, U.; Krahl, R.; Nehring, C.; Jäkel, N.; Cross, M.; Maier, J.; Niederwieser, D.; Wickenhauser, C. JAK2 p.V617F allele burden in myeloproliferative neoplasms one month after allogeneic stem cell transplantation significantly predicts outcome and risk of relapse. Haematologica 2013, 98, 722–728. [Google Scholar] [CrossRef][Green Version]
- Morley, A.A. Digital PCR: A brief history. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef]
- Nystrand, C.F.; Ghanima, W.; Waage, A.; Jonassen, C.M. JAK2 V617F mutation can be reliably detected in serum using droplet digital PCR. Int. J. Lab. Hematol. 2018, 40, 181–186. [Google Scholar] [CrossRef]
- Milbury, C.A.; Zhong, Q.; Lin JWilliams, M.; Olson, J.; Link, D.R.; Hutchison, B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol. Detect Quantif. 2014, 1, 8–22. [Google Scholar] [CrossRef]
- Tholen, D.W.; Linnet, K.; Kondratovich, M.; Armbruster, D.A.; Garrett, P.E.; Jones, R.L.; Kroll, M.H.; Lequin, R.M.; Pankratz, T.J.; Scassellati, G.A.; et al. Protocols for Determination of Limits of Detection and Limits of Quantitation; Approved Guideline; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2004; Volume 24, p. 34. Available online: https://pdfs.semanticscholar.org/9afc/36c690cfbf76f9251464045564a5ee073d7b.pdf (accessed on 5 March 2020).
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Didone, A.; Nardinelli, L.; Marchiani, M.; Ruiz, A.R.L.; de Lima Costa, A.L.; Lima, I.S.; Santos, N.M.; Sanabani, S.S.; Bendit, I. Comparative study of different methodologies to detect the JAK2 V617F mutation in chronic BCR-ABL1 negative myeloproliferative neoplasms. Pract. Lab. Med. 2016, 4, 30–37. [Google Scholar] [CrossRef]
- Lee, E.; Lee, K.J.; Park, H.; Chung, J.Y.; Lee, M.N.; Chang, M.H.; Yoo, J.; Lee, H.; Kong, S.Y.; Eom, H.S. Clinical Implications of Quantitative JAK2 V617F Analysis using Droplet Digital PCR in Myeloproliferative Neoplasms. Ann. Lab. Med. 2018, 38, 147–154. [Google Scholar] [PubMed]
| Characteristics | ET | PV | PMF | Control |
|---|---|---|---|---|
| Number of patients | 88 | 63 | 44 | 50 |
| Median age, years (range) | 66 (32–96) | 68 (37–89) | 67 (38–94) | 58 (19–66) |
| Gender, M/F | 49 F 39 M | 21 F 42 M | 18 F 26 M | 8 F 12 M |
| Median hemoglobin, g/dl (range) | 13.42 (8.1–17) | 16.1 (10.3–19.8) | 12.42 (7.8–23.2) | 14.9 (12.2–16.8) |
| Median hematocrit, % (range) | 41.65 (24.4–52) | 50.4 (33.7–63) | 38.68 (22.6–73.9) | 43.6 (26–61.2) |
| Median red blood cell, M/µL (range) | 5.02 (3.29–6.1) | 6.23 (3.39–8.96) | 6.16 (2.88–50.05) | 5.3 (4.19–5.3) |
| Median white blood cell, K/µL (range) | 8.1238 (2.9–14.6) | 10.46 (4.82–46.96) | 16.22 (1.46–70.91) | 7.87 (4.9–11.6) |
| Median platelets, K/µL (range) | 651.73 (93–1352) | 434.08 (108–879) | 387.03 (15–1492) | 490.3 (340–790) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Rocca, F.; Grieco, V.; Ruggieri, V.; Zifarone, E.; Villani, O.; Zoppoli, P.; Russi, S.; Laurino, S.; Falco, G.; Calice, G.; et al. Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2 V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study. Diagnostics 2020, 10, 143. https://doi.org/10.3390/diagnostics10030143
La Rocca F, Grieco V, Ruggieri V, Zifarone E, Villani O, Zoppoli P, Russi S, Laurino S, Falco G, Calice G, et al. Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2 V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study. Diagnostics. 2020; 10(3):143. https://doi.org/10.3390/diagnostics10030143
Chicago/Turabian StyleLa Rocca, Francesco, Vitina Grieco, Vitalba Ruggieri, Emanuela Zifarone, Oreste Villani, Pietro Zoppoli, Sabino Russi, Simona Laurino, Geppino Falco, Giovanni Calice, and et al. 2020. "Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2 V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study" Diagnostics 10, no. 3: 143. https://doi.org/10.3390/diagnostics10030143
APA StyleLa Rocca, F., Grieco, V., Ruggieri, V., Zifarone, E., Villani, O., Zoppoli, P., Russi, S., Laurino, S., Falco, G., Calice, G., Marinaccio, A., Natalicchio, M. I., Albano, F., & Musto, P. (2020). Superiority of Droplet Digital PCR Over Real-Time Quantitative PCR for JAK2 V617F Allele Mutational Burden Assessment in Myeloproliferative Neoplasms: A Retrospective Study. Diagnostics, 10(3), 143. https://doi.org/10.3390/diagnostics10030143






