Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer
Abstract
1. Introduction
2. Results
2.1. Demographics of the Study Groups
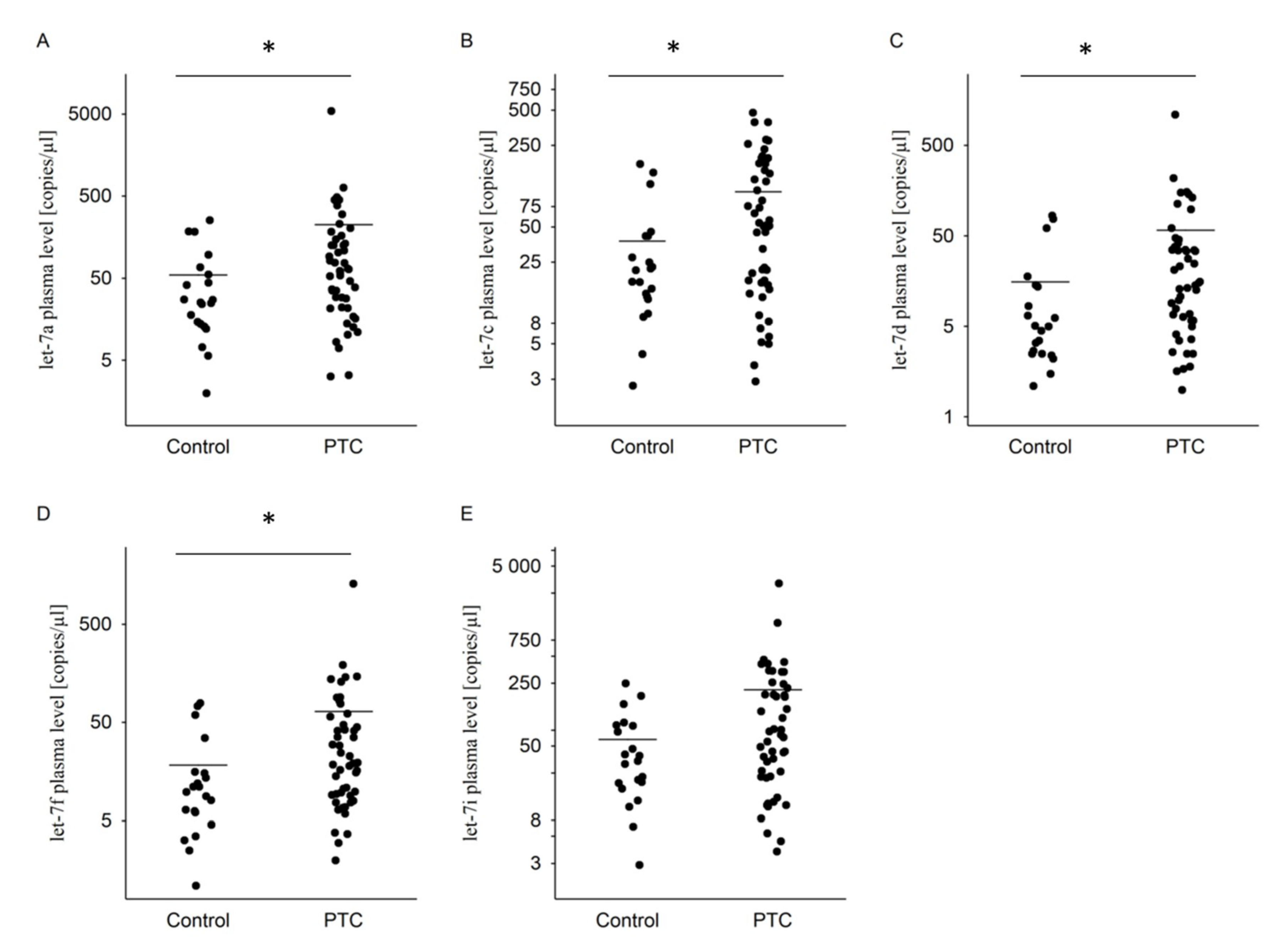
2.2. Plasma miRNA in PTC and Healthy Control Groups
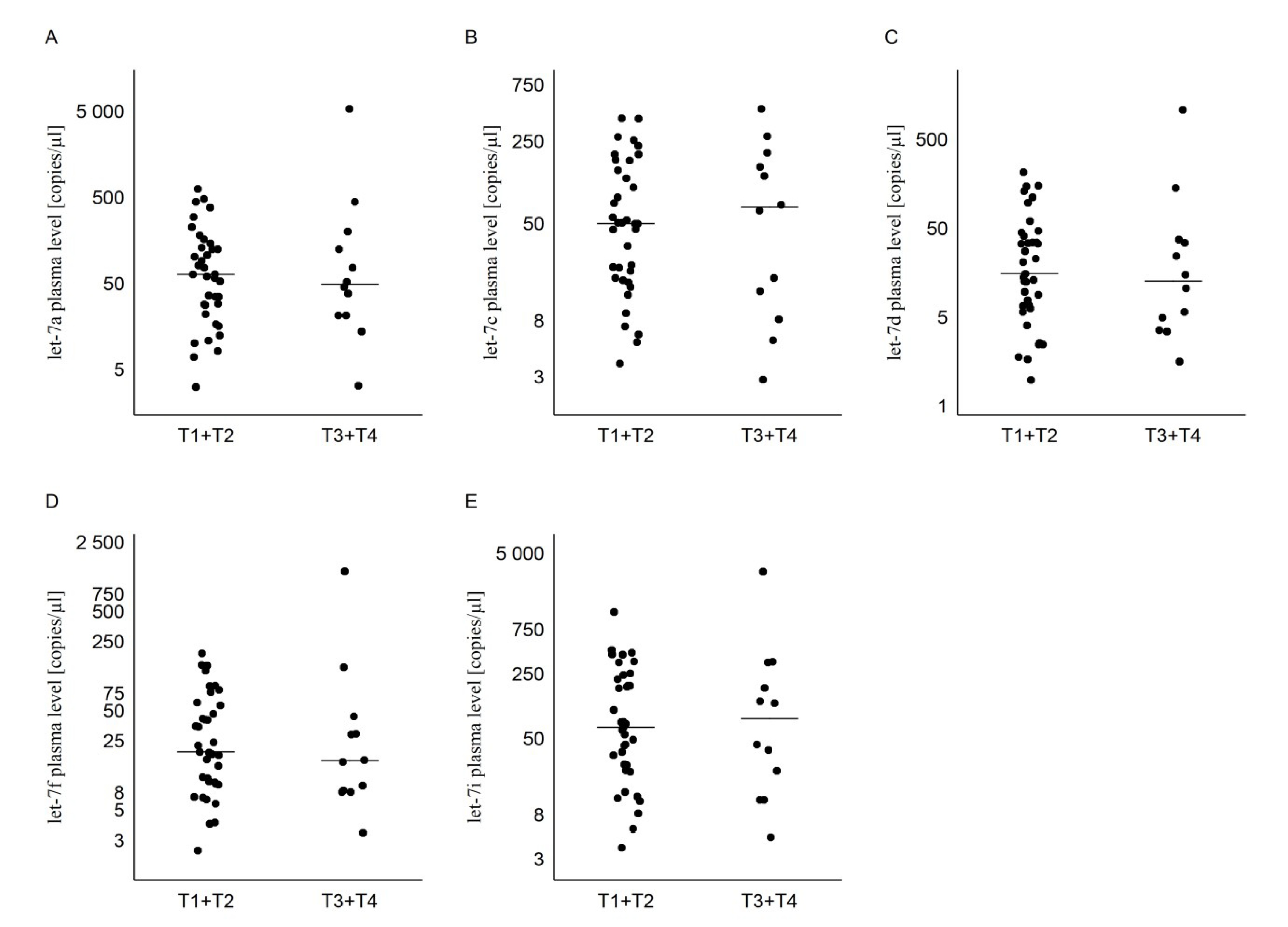
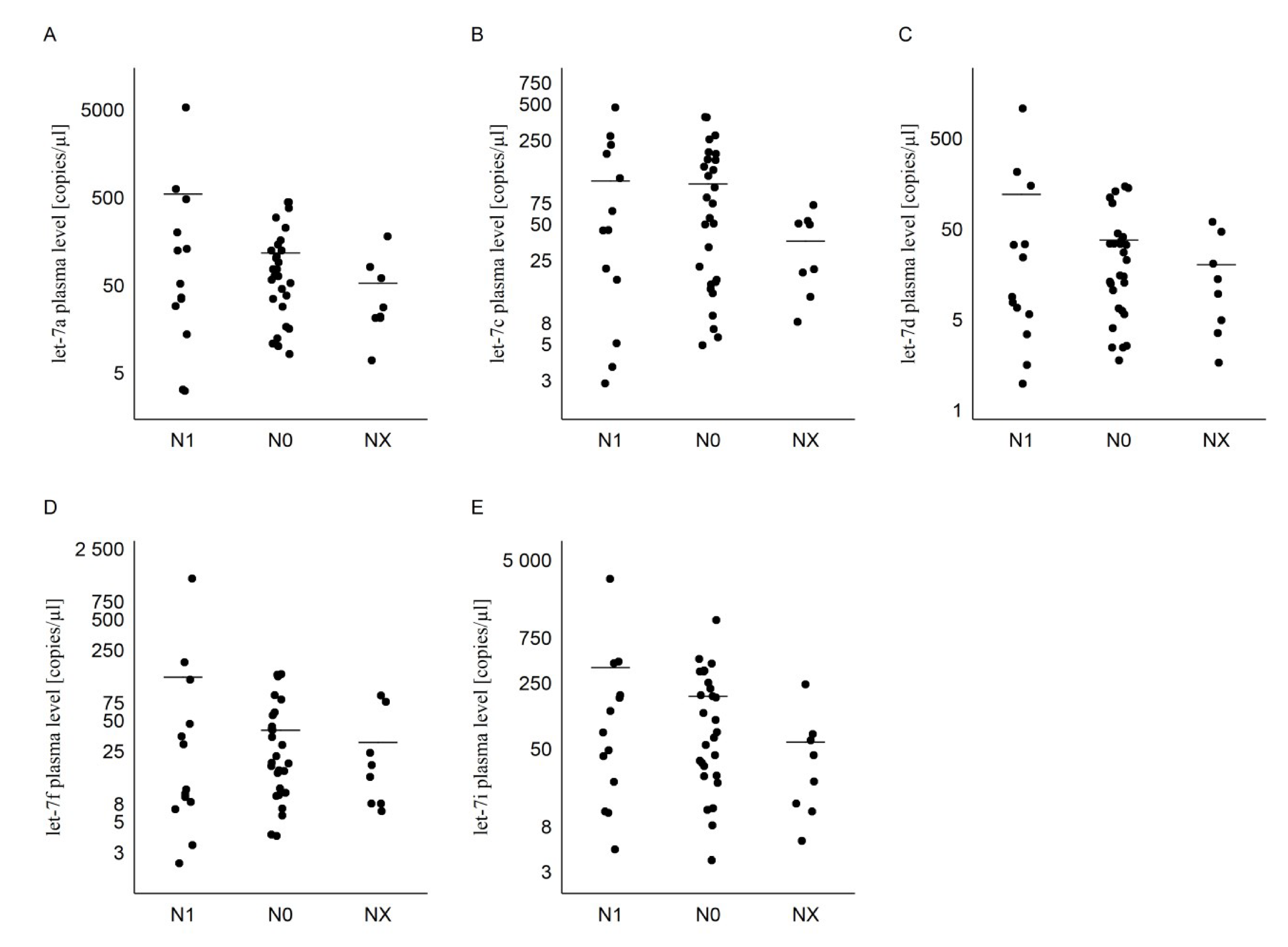
2.3. Tumor and Lymph Nodes Characteristics in PTC Group
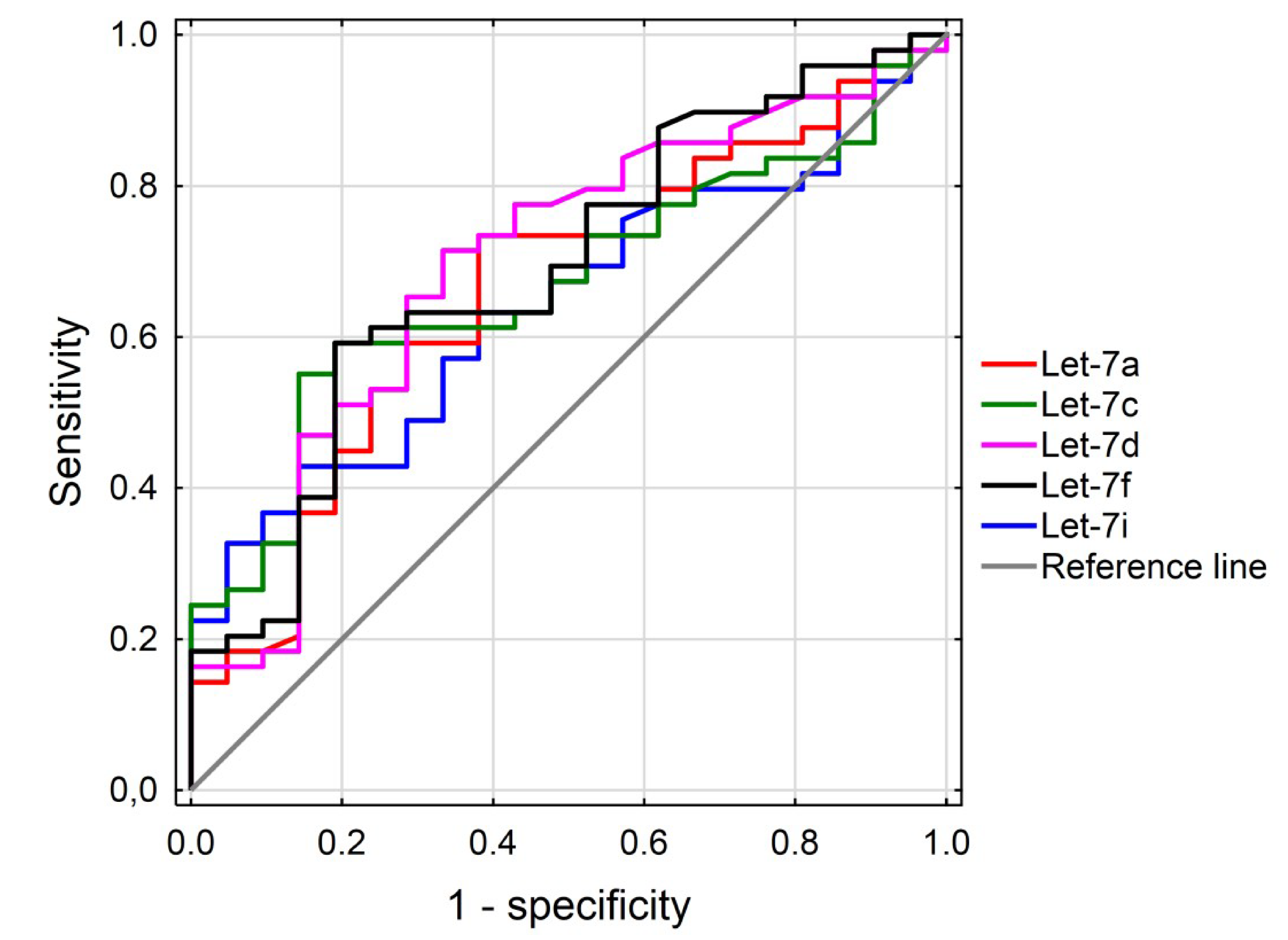
2.4. Receiver Operating Characteristic (ROC) Curve
3. Discussion
4. Materials and Methods
4.1. Patients and Blood Samples
4.2. Blood Sample Processing and miRNA Extraction
4.3. Reverse Transcription
4.4. Digital Droplet PCR
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AUC | areas under the curve |
| ddPCR | droplet digital PCR |
| FNAB | fine needle aspiration biopsy |
| HDL | high density lipoprotein |
| miRNAs | microRNAs |
| Npm1 | nucleophosmin 1 |
| PTC | papillary thyroid cancer |
| qPCR | quantitative PCR |
| ROC | receiver operating characteristic |
| TAMs | Tumor associated macrophages |
References
- Vaccarella, S.; Franceschi, S.; Bray, F.; Wild, C.P.; Plummer, M.; Dal Maso, L. Worldwide Thyroid-Cancer Epidemic? The Increasing Impact of Overdiagnosis. N. Engl. J. Med. 2016, 375, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.; Wintheiser, G.; Wolfe, K.M.; Droessler, J.; Silberstein, P.T. Epidemiology of Thyroid Cancer: A Review of the National Cancer Database, 2000–2013. Cureus 2019, 11, e4127. [Google Scholar] [CrossRef] [PubMed]
- Witt, R.L.; Ferris, R.L.; Pribitkin, E.A.; Sherman, S.I.; Steward, D.L.; Nikiforov, Y.E. Diagnosis and management of differentiated thyroid cancer using molecular biology. Laryngoscope 2013, 123, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Perdas, E.; Stawski, R.; Kaczka, K.; Nowak, D.; Zubrzycka, M. Altered levels of circulating nuclear and mitochondrial DNA in patients with Papillary Thyroid Cancer. Sci. Rep. 2019, 9, 14438. [Google Scholar] [CrossRef]
- Perdas, E.; Stawski, R.; Nowak, D.; Zubrzycka, M. Potential of Liquid Biopsy in Papillary Thyroid Carcinoma in Context of miRNA, BRAF and p53 Mutation. Curr. Drug Targets 2018, 19, 1721–1729. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef]
- He, Y.; Lin, J.; Kong, D.; Huang, M.; Xu, C.; Kim, T.K.; Etheridge, A.; Luo, Y.; Ding, Y.; Wang, K. Current state of circulating microRNAs as cancer biomarkers. Clin. Chem. 2015, 61, 1138–1155. [Google Scholar] [CrossRef]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef]
- Perdas, E.; Stawski, R.; Nowak, D.; Zubrzycka, M. The role of miRNA in papillary thyroid cancer in the context of miRNA let-7 family. Int. J. Mol. Sci. 2016, 17, 909. [Google Scholar] [CrossRef]
- Campomenosi, P.; Gini, E.; Noonan, D.M.; Poli, A.; D’Antona, P.; Rotolo, N.; Dominioni, L.; Imperatori, A. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 2016, 16, 60. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Kelly, R.; Newell, J.; Kerin, M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 2010, 15, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Kelly, B.; Miller, N.; Sweeney, K.; Durkan, G.; Rogers, E.; Walsh, K.; Kerin, M. A Circulating MicroRNA Signature as a Biomarker for Prostate Cancer in a High Risk Group. J. Clin. Med. 2015, 4, 1369–1379. [Google Scholar] [CrossRef]
- Tsujiura, M.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Takeshita, H.; Kosuga, T.; Konishi, H.; Morimura, R.; Deguchi, K.; Fujiwara, H.; et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 2010, 102, 1174–1179. [Google Scholar] [CrossRef]
- Ghanbari, R.; Mosakhani, N.; Sarhadi, V.; Armengol, G.; Nouraee, N.; Mohammadkhani, A.; Khorrami, S.; Arefian, E.; Paryan, M.; Malekzadeh, R.; et al. Simultaneous Underexpression of let-7a-5p and let-7f-5p microRNAs in Plasma and Stool Samples from Early Stage Colorectal Carcinoma. Biomark. Cancer 2015, 2015, 39–48. [Google Scholar] [CrossRef]
- Khalighfard, S.; Alizadeh, A.M.; Irani, S.; Omranipour, R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci. Rep. 2018, 8, 17981. [Google Scholar] [CrossRef]
- Li, X.-X.; Gao, S.-Y.; Wang, P.-Y.; Zhou, X.; Li, Y.-J.; Yu, Y.; Yan, Y.-F.; Zhang, H.-H.; Lv, C.-J.; Zhou, H.-H.; et al. Reduced expression levels of let-7c in human breast cancer patients. Oncol. Lett. 2015, 9, 1207–1212. [Google Scholar] [CrossRef]
- Qattan, A.; Intabli, H.; Alkhayal, W.; Eltabache, C.; Tweigieri, T.; Amer, S. Bin Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 2017, 17, 799. [Google Scholar] [CrossRef]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol. Cancer 2019, 18, 76. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-J.; Xu, Q.; Sun, L.-P.; Dong, Q.-G.; He, C.; Yuan, Y. Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease. Tumor Biol. 2015, 36, 3337–3343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Shan, H.; Su, Y.; Xia, K.; Zou, R.; Shao, Q. Let-7a inhibits migration, invasion and tumor growth by targeting AKT2 in papillary thyroid carcinoma. Oncotarget 2017, 8, 69746–69755. [Google Scholar] [CrossRef] [PubMed]
- Elise, M.; Graham, R.; Hart, R.D.; Douglas, S.; Makki, F.M.; Pinto, D.; Butler, A.L.; Bullock, M.; Rigby, M.H.; Trites, J.R.B.; et al. Serum microRNA profiling to distinguish papillary thyroid cancer from benign thyroid masses. J. Otolaryngol. Head Neck Surg. 2015, 44, 33. [Google Scholar]
- Damanakis, A.I.; Eckhardt, S.; Wunderlich, A.; Roth, S.; Wissniowski, T.T.; Bartsch, D.K.; Di Fazio, P. MicroRNAs let7 expression in thyroid cancer: Correlation with their deputed targets HMGA2 and SLC5A5. J. Cancer Res. Clin. Oncol. 2016, 142, 1213–1220. [Google Scholar] [CrossRef]
- Li, H.; Zhao, L.; Zhang, Z.; Zhang, H.; Ding, C.; Su, Z. Roles of microRNA let-7b in papillary thyroid carcinoma by regulating HMGA2. Tumor Biol. 2017, 39, 1–10. [Google Scholar] [CrossRef]
- Braun, J.; Hoang-Vu, C.; Dralle, H.; Hüttelmaier, S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010, 29, 4237–4244. [Google Scholar] [CrossRef]
- Swierniak, M.; Wojcicka, A.; Czetwertynska, M.; Stachlewska, E.; Maciag, M.; Wiechno, W.; Gornicka, B.; Bogdanska, M.; Koperski, L.; De La Chapelle, A.; et al. In-depth characterization of the MicroRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1401–E1409. [Google Scholar] [CrossRef]
- Yu, S.; Liu, Y.; Wang, J.; Guo, Z.; Zhang, Q.; Yu, F.; Zhang, Y.; Huang, K.; Li, Y.; Song, E.; et al. Circulating MicroRNA Profiles as Potential Biomarkers for Diagnosis of Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, 2084–2092. [Google Scholar] [CrossRef]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 2006, 13, 497–508. [Google Scholar] [CrossRef]
- Li, M.; Song, Q.; Li, H.; Lou, Y.; Wang, L. Circulating miR-25-3p and miR-451a may be potential biomarkers for the diagnosis of papillary thyroid carcinoma. PLoS ONE 2015, 10, e0132403. [Google Scholar]
- de Vasconcellos, J.F.; Byrnes, C.; Lee, Y.T.; Allwardt, J.M.; Kaushal, M.; Rabel, A.; Miller, J.L. Tough decoy targeting of predominant let-7 miRNA species in adult human hematopoietic cells. J. Transl. Med. 2017, 15, 169. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Q.; Yao, J.; Jiang, H.; Xiao, C.; Wu, F. MicroRNA let-7g and let-7i inhibit hepatoma cell growth concurrently via downregulation of the anti-apoptoticprotein B-cell lymphoma-extra large. Oncol. Lett. 2015, 9, 213–218. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jin, B.; Wang, W.; Meng, X.; Du, G.; Li, J.; Zhang, S.; Zhou, B.; Fu, Z. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer 2016, 16, 863. [Google Scholar] [CrossRef] [PubMed]
- Baer, C.; Squadrito, M.L.; Laoui, D.; Thompson, D.; Hansen, S.K.; Kiialainen, A.; Hoves, S.; Ries, C.H.; Ooi, C.H.; De Palma, M. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 2016, 18, 790–802. [Google Scholar] [CrossRef]
- Pobezinsky, L.A.; Wells, A.C. Let’s fight cancer: Let-7 is a tool to enhance antitumor immune responses. Futur. Oncol. 2018, 14, 1141–1145. [Google Scholar] [CrossRef]
- Careccia, S.; Mainardi, S.; Pelosi, A.; Gurtner, A.; Diverio, D.; Riccioni, R.; Testa, U.; Pelosi, E.; Piaggio, G.; Sacchi, A.; et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene 2009, 28, 4034–4040. [Google Scholar] [CrossRef]
- Sorrentino, A.; Liu, C.G.; Addario, A.; Peschle, C.; Scambia, G.; Ferlini, C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol. Oncol. 2008, 111, 478–486. [Google Scholar] [CrossRef]
- Yin, J.; Zhao, J.; Hu, W.; Yang, G.; Yu, H.; Wang, R.; Wang, L.; Zhang, G.; Fu, W.; Dai, L.; et al. Disturbance of the let-7/LIN28 double-negative feedback loop is associated with radio- and chemo-resistance in non-small cell lung cancer. PLoS ONE 2017, 12, e0172787. [Google Scholar] [CrossRef]
- Xiao, M.; Cai, J.; Cai, L.; Jia, J.; Xie, L.; Zhu, Y.; Huang, B.; Jin, D.; Wang, Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J. Ovarian Res. 2017, 10, 24. [Google Scholar] [CrossRef]
- Cai, J.; Yang, C.; Yang, Q.; Ding, H.; Jia, J.; Guo, J.; Wang, J.; Wang, Z. Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis 2013, 2, e75. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Kaur, S.; Volinia, S.; Greshock, J.; Lassus, H.; Hasegawa, K.; Liang, S.; Leminen, A.; Deng, S.; Smith, L.; et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008, 68, 10307–10314. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, X.; Hu, J.; Lu, Q. Targeted regulation of MiR-98 on E2F1 increases chemosensitivity of leukemia cells K562/A02. Onco. Targets. Ther. 2017, 10, 3233–3239. [Google Scholar] [CrossRef] [PubMed]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef]
- Jarząb, B.; Dedecjus, M.; Handkiewicz-Junak, D.; Lange, D.; Lewiński, A.; Nasierowska-Guttmejer, A.; Ruchała, M.; Słowińska-Klencka, D.; Nauman, J. Diagnostyka i leczenie raka tarczycy. Endokrynol. Pol. 2016, 67, 74–145. [Google Scholar] [CrossRef]
- Tuck, M.; Turgeon, D.K.; Brenner, D.E. Serum and Plasma Collection: Preanalytical Variables and Standard Operating Procedures in Biomarker Research. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Elsevier Inc: Amsterdam, The Netherlands, 2013; pp. 77–85. ISBN 9780123944467. [Google Scholar]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef]
- Ma, J.; Li, N.; Guarnera, M.; Jiang, F. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomark. Insights 2013, 8, 127–136. [Google Scholar] [CrossRef]
| Variability | Control | PTC |
|---|---|---|
| Gender | 21 | 49 |
| Male | 9 (43%) | 5 (10%) |
| Female | 12 (57%) | 44 (90%) |
| Age (years) | ||
| Median | 47 | 48 |
| Range | 29–67 | 20–76 |
| T&N staging | ||
| T1 + T2 | — | 37 (76%) |
| T3 + T4 | — | 12 (24%) |
| N0 | — | 28 (57%) |
| N1 | — | 13 (27%) |
| NX | — | 8 (16%) |
| miRNAs | Control (Copies/µL) n = 21 | PTC (Copies/µL) n = 49 |
|---|---|---|
| Let-7a | 55 ± 69 (25) | 224 ± 776 (62) * |
| Let-7c | 38 ± 47 (21) | 100 ± 115 (51) * |
| Let-7d | 16 ± 25 (5) | 58 ± 159 (15) * |
| Let-7f | 18 ± 23 (10) | 65 ± 185 (19) * |
| Let-7i | 59 ± 64 (34) | 212 ± 483 (67) |
| PTC (Copies/µL) | |||||
| Variability | Let-7a | Let-7c | Let-7d | Let-7f | Let-7i |
| Gender | |||||
| Male | 38 ± 17 (37) | 32 ± 27(20) | 10 ± 7 (9) | 13 ± 7 (10) | 48 ± 27 (44) |
| Female | 245 ± 817 (72) | 107 ± 119 (53) | 63 ± 167 (19) | 71 ± 194 (21) | 230 ± 507 (72) |
| T&N staging | |||||
| T1 + T2 | 121 ± 150 (65) | 93 ± 106 (51) | 39 ± 52 (16) | 42 ± 47 (20) | 160 ± 222 (67) |
| T3 + T4 | 543 ± 1 551 (50) | 119 ± 143 (69) | 115 ± 311 (13) | 134 ± 367 (16) | 372 ± 905 (83) |
| N0 | 119 ± 130 (72) | 110 ± 114 (67) | 38 ± 45 (19) | 41 ± 43 (21) | 183 ± 240 (90) |
| N1 | 555 ± 1 484 (53) | 116 ± 142 (45) | 123 ± 300 (9) | 137 ± 352 (11) | 367 ± 869 (76) |
| NX | 54 ± 59 (25) | 36 ± 24 (36) | 20 ± 22 (12) | 31 ± 33 (16) | 60 ± 79 (33) |
| Control(Copies/µL) | |||||
| Let-7a | Let-7c | Let-7d | Let-7f | Let-7i | |
| Gender | |||||
| Male | 30 ± 28 (25) | 18 ± 13 (21) | 5 ± 4 (5) | 11 ± 10 (10) | 30 ± 22 (32) |
| Female | 73 ± 86 (33) | 53 ± 58 (20) | 24 ± 31 (7) | 24 ± 29(10) | 81 ± 77 (61) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perdas, E.; Stawski, R.; Kaczka, K.; Zubrzycka, M. Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer. Diagnostics 2020, 10, 130. https://doi.org/10.3390/diagnostics10030130
Perdas E, Stawski R, Kaczka K, Zubrzycka M. Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer. Diagnostics. 2020; 10(3):130. https://doi.org/10.3390/diagnostics10030130
Chicago/Turabian StylePerdas, Ewelina, Robert Stawski, Krzysztof Kaczka, and Maria Zubrzycka. 2020. "Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer" Diagnostics 10, no. 3: 130. https://doi.org/10.3390/diagnostics10030130
APA StylePerdas, E., Stawski, R., Kaczka, K., & Zubrzycka, M. (2020). Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer. Diagnostics, 10(3), 130. https://doi.org/10.3390/diagnostics10030130






