Identification of Key Biomarkers in Bladder Cancer: Evidence from a Bioinformatics Analysis
Abstract
1. Introduction
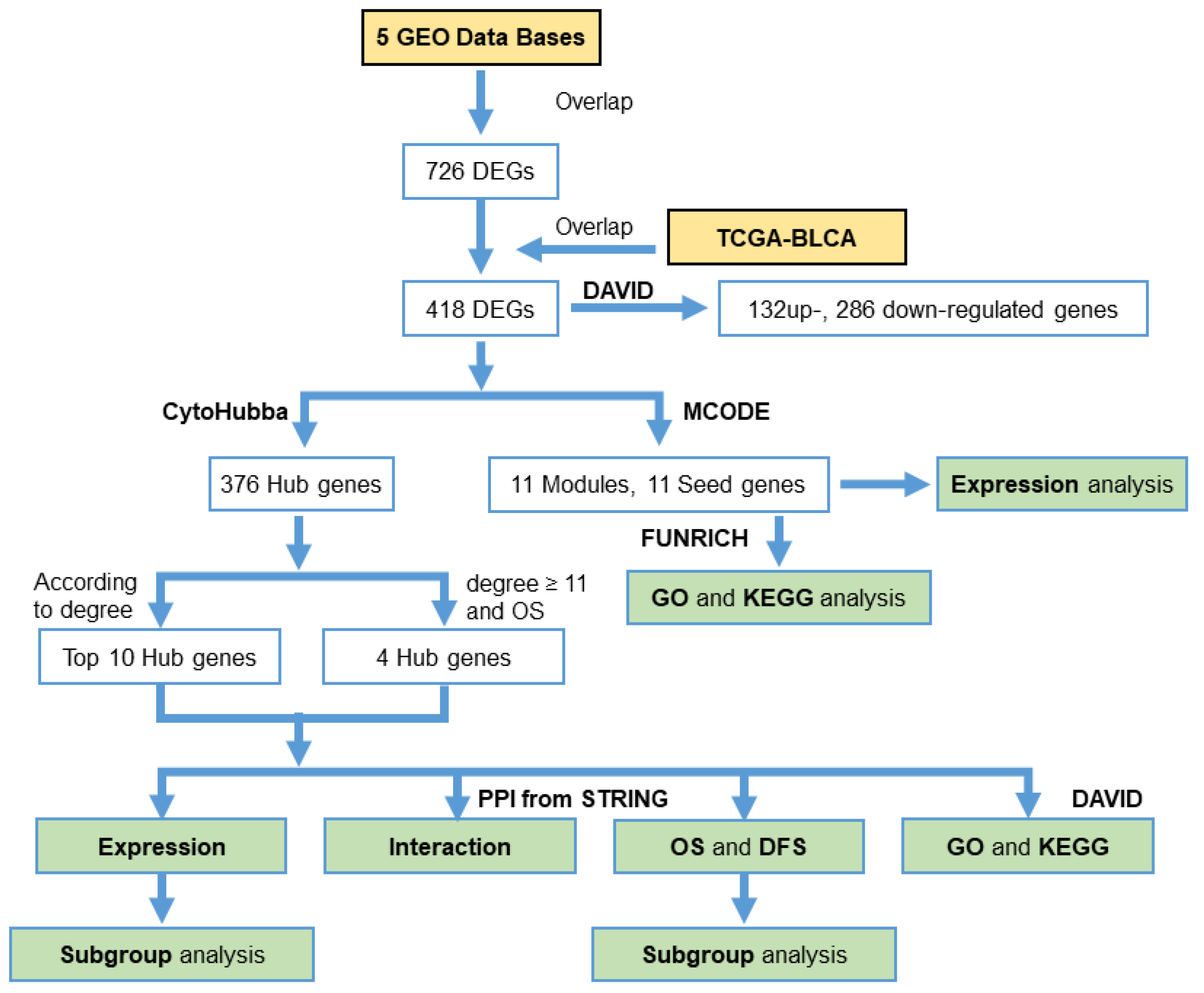
2. Materials and Methods
2.1. Acquisition of Microarray Data
2.2. Identification of DEGs
2.3. GO and KEGG Analyses of DEGs
2.4. Protein‒Protein Interaction (PPI) Network Analysis
2.5. Clinical Data Analysis and Oncomine Analysis
3. Results
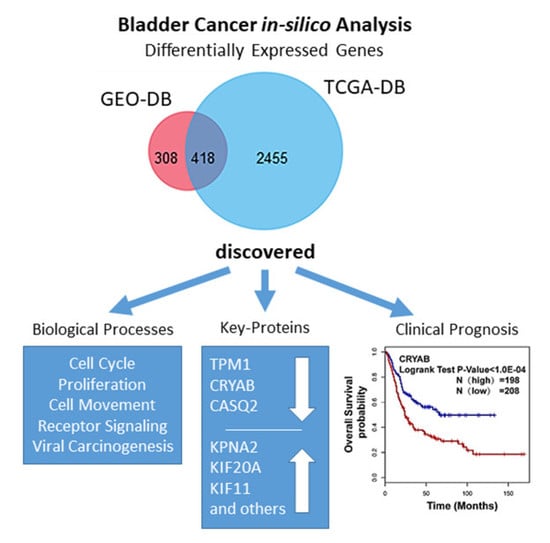
3.1. Identification of DEGs in BCa
3.2. GO and KEGG Enrichment Analyses of DEGs
3.2.1. GO Biological Process (BP)
3.2.2. GO Cellular Component (CC)
3.2.3. GO Molecular Function (MF)
3.2.4. KEGG Pathway Analysis
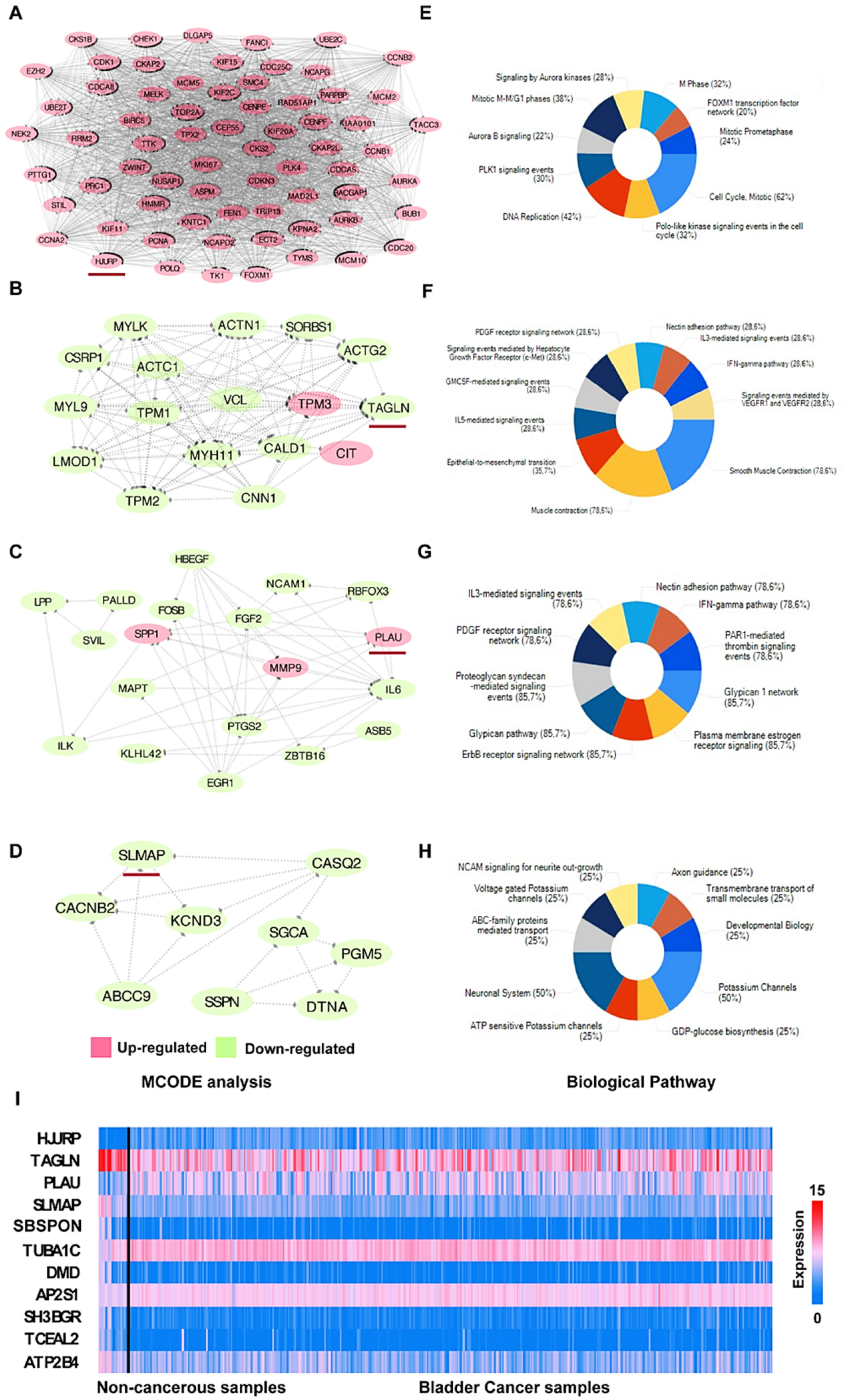
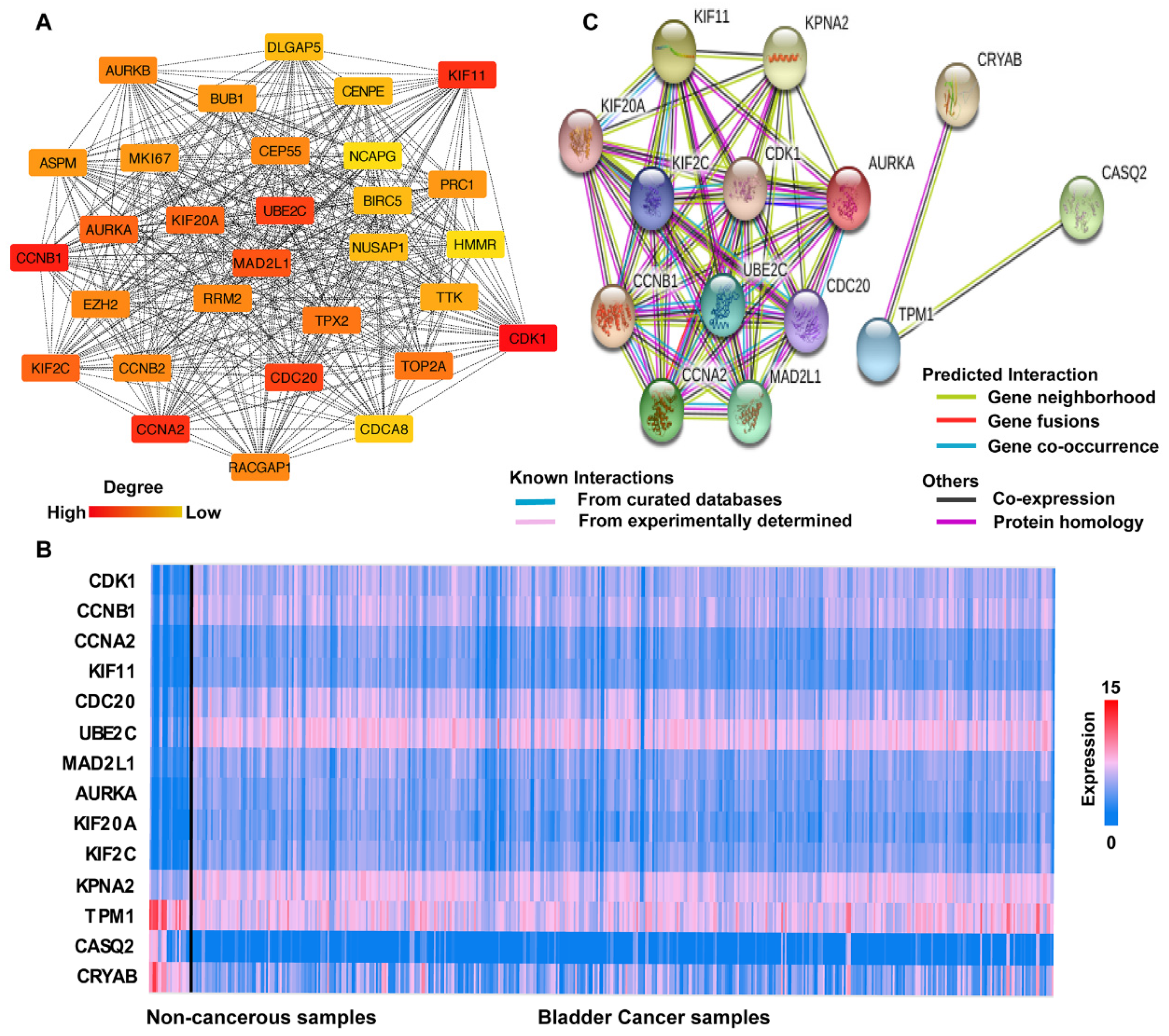
3.3. PPI Network Analysis
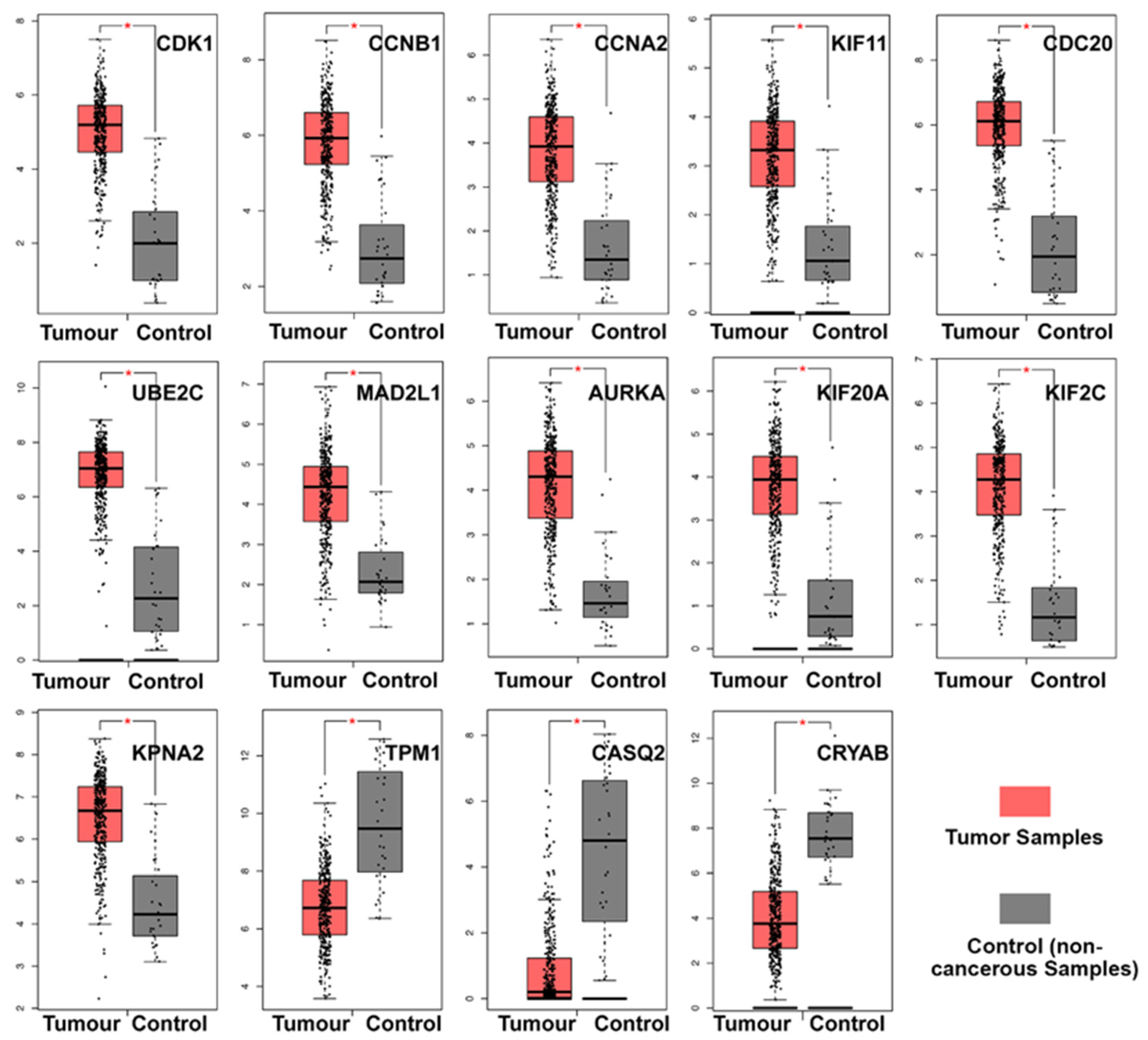
3.4. Hub Gene Selection and Analysis
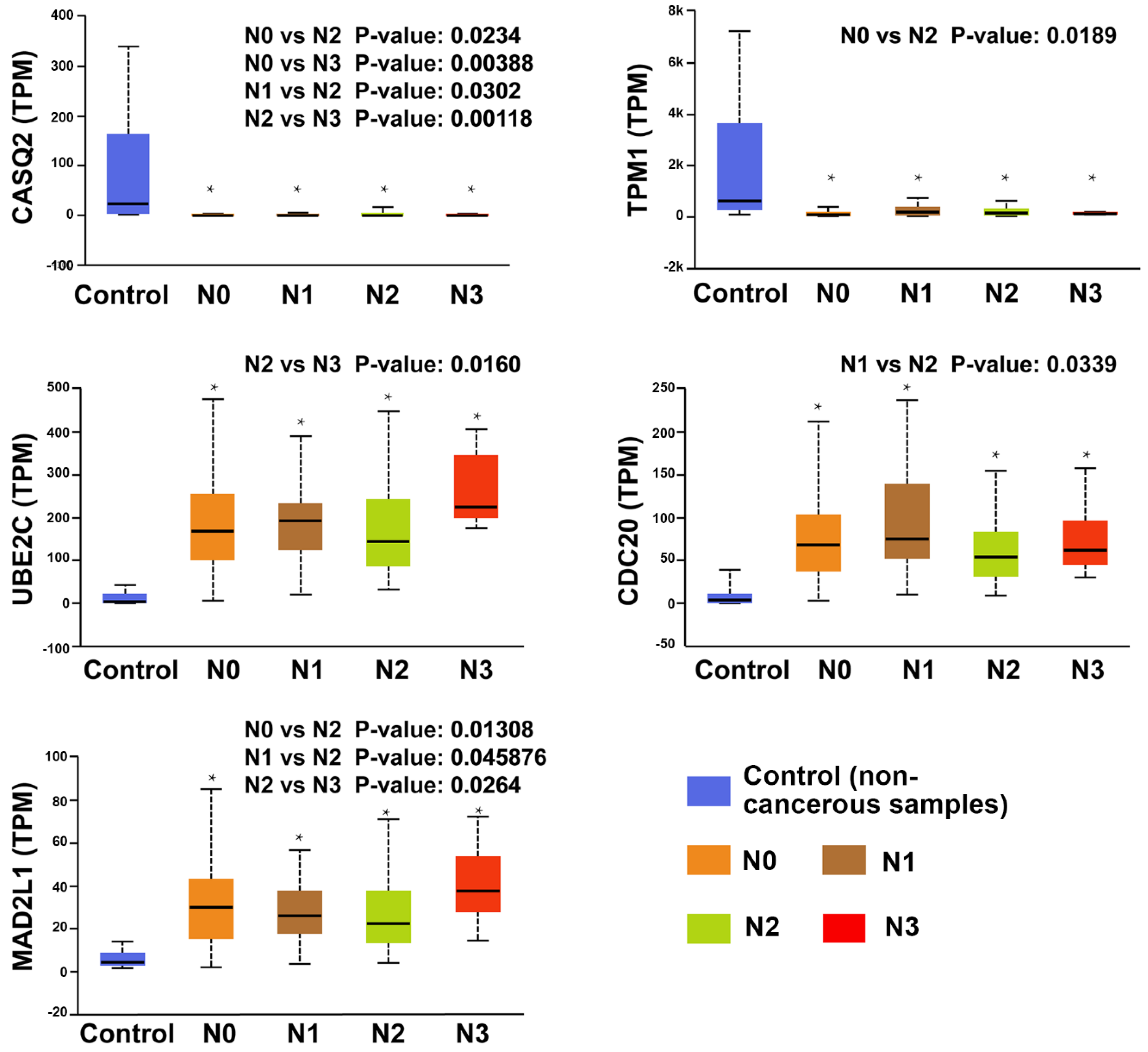
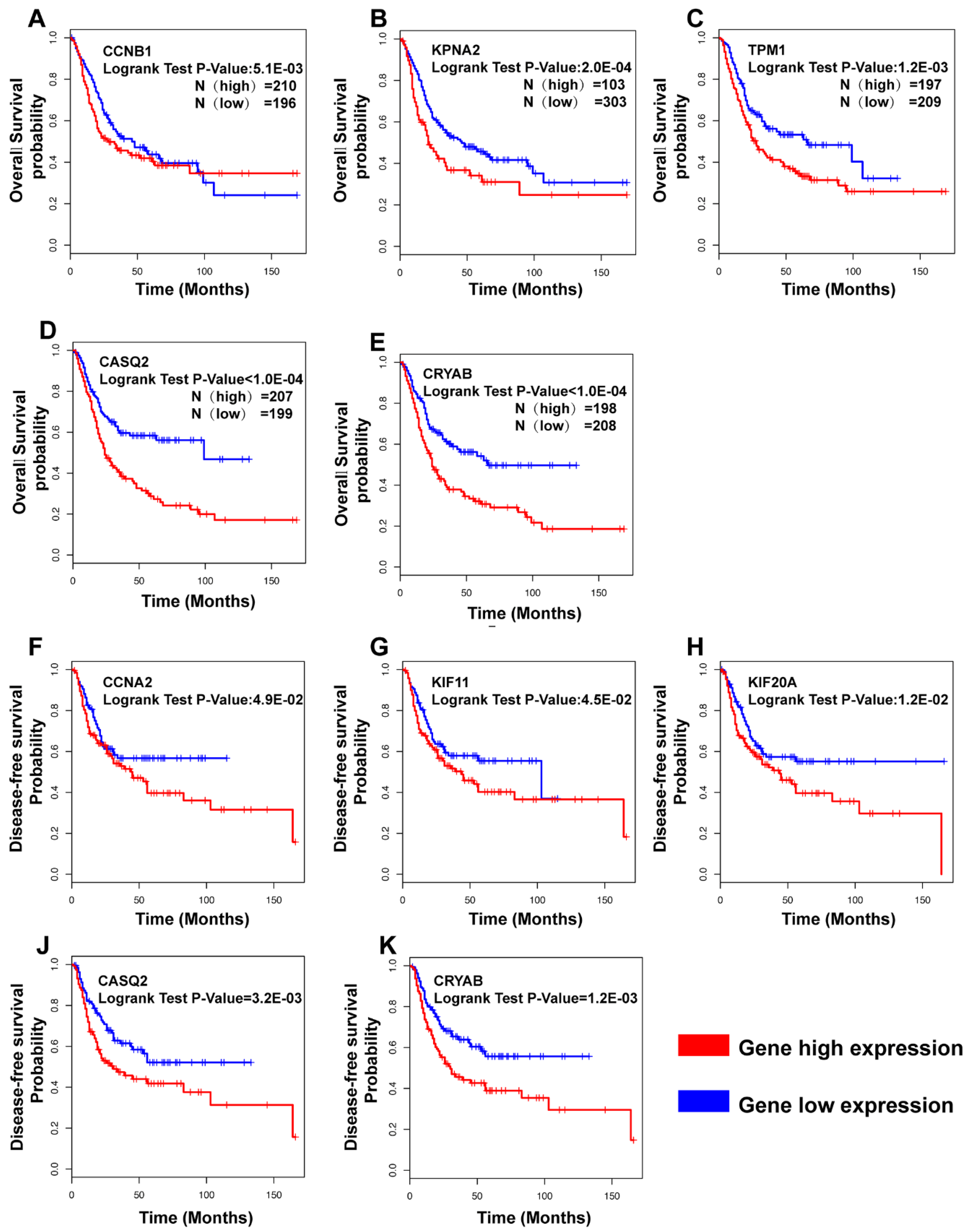
3.5. Clinical Analysis and Oncomine Analysis Outcomes of Hub Genes
4. Discussion
4.1. Biological Progression Potentially Associated with BCa
4.2. The Hub Genes Are Potentially Associated with the Clinical Outcomes
4.3. The Hub Genes Potentially Associated with the Clinical Prognosis in Previous Research
4.4. Major Findings of the Recent Study
- (i)
- TPM1, CRYAB, and CASQ2 were significantly downregulated in BCa, while the remaining 11 hub genes were significantly upregulated.
- (ii)
- All hub genes showed a significant difference between the lymph node metastasis status and noncancerous tissues. In particular, UBE2C, CDC20, MAD2L1, TPM1, and CASQ2 potentially play a pivotal role in lymph node metastasis.
- (iii)
- CCNB1, KPNA2, TPM1, CASQ2, and CRYAB were correlated with prognosis in overall survival (OS) analysis, while CCNA2, KIF11, KIF20A, CASQ2, and CRYAB were correlated with disease-free survival (DFS). Interestingly, CRYAB and CASQ2 were correlated with both OS and DFS.
- (iv)
- We here report evidence for an up to now unrecognized possible role of CASQ2 in cancer and for CRYAB in bladder cancer. In combination with the results from the HPA, our study suggests that CRYAB would be a candidate biomarker in BCa, followed by CASQ2, TPM1, and KPNA2.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BCa | Bladder cancer |
| GEO | Gene Expression Omnibus |
| TCGA-BLCA | Cancer Genome Atlas Bladder Cancer |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | Protein‒protein interaction |
| FC | Fold change |
| RLE | Relative log expression |
| OS | Overall survival |
| DFS | Disease-free survival |
| HJURP | Holliday junction recognition protein |
| TAGLN | Transgelin |
| PLAU | Urokinase |
| SLMAP | Sarcolemmal membrane-associated protein |
| NMIBC | Non-muscle-invasive bladder cancer |
| MIBC | Muscle-invasive bladder cancer |
| DEGs | Differentially expressed genes |
| MCODE | Molecular complex detection |
| GEPIA | Gene expression profiling interactive analysis |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
| cGMP-PKG | Cyclic guanosine monophosphate-dependent protein kinase |
| cAMP | Cyclic adenosine monophosphate |
| FPKM | Fragments per kilobase million |
| TPM | Transcripts per million |
References
- Kulkarni, G.S.; Black, P.C.; Sridhar, S.S.; Kapoor, A.; Zlotta, A.R.; Shayegan, B.; Rendon, R.A.; Chung, P.; van der Kwast, T.; Alimohamed, N.; et al. Canadian Urological Association guideline: Muscle-invasive bladder cancer. Can. Urol. Assoc. J. 2019, 13, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Flaig, T.W.; Spiess, P.E.; Agarwal, N.; Bangs, R.; Boorjian, S.A.; Buyyounouski, M.K.; Downs, T.M.; Efstathiou, J.A.; Friedlander, T.; Greenberg, R.E.; et al. NCCN Guidelines Insights: Bladder Cancer, Version 5.2018. J. Natl. Compr. Cancer Netw. JNCCN 2018, 16, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Comperat, E.M.; Gontero, P.; Mostafid, A.H.; Palou, J.; van Rhijn, B.W.G.; Roupret, M.; Shariat, S.F.; Sylvester, R.; et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ)—2019 Update. Eur. Urol. 2019, 76, 639–657. [Google Scholar] [CrossRef] [PubMed]
- Zaghloul, M.S.; Christodouleas, J.P.; Smith, A.; Abdallah, A.; William, H.; Khaled, H.M.; Hwang, W.T.; Baumann, B.C. Adjuvant Sandwich Chemotherapy Plus Radiotherapy vs. Adjuvant Chemotherapy Alone for Locally Advanced Bladder Cancer After Radical Cystectomy: A Randomized Phase 2 Trial. JAMA Surg. 2018, 153, e174591. [Google Scholar] [CrossRef] [PubMed]
- Czerniak, B.; Dinney, C.; McConkey, D. Origins of Bladder Cancer. Annu. Rev. Pathol. 2016, 11, 149–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.T.; Pak, J.; Shapiro, E.; Sun, T.T.; Wu, X.R. Urothelium-specific expression of an oncogene in transgenic mice induced the formation of carcinoma in situ and invasive transitional cell carcinoma. Cancer Res. 1999, 59, 3512–3517. [Google Scholar] [PubMed]
- Zhang, Z.T.; Pak, J.; Huang, H.Y.; Shapiro, E.; Sun, T.T.; Pellicer, A.; Wu, X.R. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene 2001, 20, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Patel, R.; Liu, Y.; Singh, L.B.; Taketo, M.M.; Wu, X.R.; Leung, H.Y.; Sansom, O.J. Ras mutation cooperates with beta-catenin activation to drive bladder tumourigenesis. Cell Death Dis. 2011, 2, e124. [Google Scholar] [CrossRef]
- Sjodahl, G.; Eriksson, P.; Patschan, O.; Marzouka, N.A.; Jakobsson, L.; Bernardo, C.; Lovgren, K.; Chebil, G.; Zwarthoff, E.; Liedberg, F.; et al. Molecular changes during progression from nonmuscle invasive to advanced urothelial carcinoma. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Wang, J.P.; Jiao, Y.; Wang, C.Y.; Xu, Z.B.; Zhang, B. Rb knockdown accelerates bladder cancer progression through E2F3 activation. Int. J. Oncol. 2017, 50, 149–160. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Wang, J.Y. Targeting the RB-pathway in cancer therapy. Clin. Cancer Res. 2010, 16, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Power, N.E.; Izawa, J. Comparison of Guidelines on Non-Muscle Invasive Bladder Cancer (EAU, CUA, AUA, NCCN, NICE). Bladder Cancer 2016, 2, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Boorjian, S.A.; Chou, R.; Clark, P.E.; Daneshmand, S.; Konety, B.R.; Pruthi, R.; Quale, D.Z.; Ritch, C.R.; Seigne, J.D.; et al. Diagnosis and Treatment of Non-Muscle Invasive Bladder Cancer: AUA/SUO Guideline. J. Urol. 2016, 196, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Alfred Witjes, J.; Lebret, T.; Comperat, E.M.; Cowan, N.C.; De Santis, M.; Bruins, H.M.; Hernandez, V.; Espinos, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef]
- Li, L.; Lei, Q.; Zhang, S.; Kong, L.; Qin, B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol. Rep. 2017, 38, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Zaravinos, A.; Lambrou, G.I.; Boulalas, I.; Delakas, D.; Spandidos, D.A. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE 2011, 6, e18135. [Google Scholar] [CrossRef]
- Zaravinos, A.; Lambrou, G.I.; Volanis, D.; Delakas, D.; Spandidos, D.A. Spotlight on differentially expressed genes in urinary bladder cancer. PLoS ONE 2011, 6, e18255. [Google Scholar] [CrossRef]
- Borisov, N.; Tkachev, V.; Suntsova, M.; Kovalchuk, O.; Zhavoronkov, A.; Muchnik, I.; Buzdin, A. A method of gene expression data transfer from cell lines to cancer patients for machine-learning prediction of drug efficiency. Cell Cycle 2018, 17, 486–491. [Google Scholar] [CrossRef]
- Zhao, F.; Lin, T.; He, W.; Han, J.; Zhu, D.; Hu, K.; Li, W.; Zheng, Z.; Huang, J.; Xie, W. Knockdown of a novel lincRNA AATBC suppresses proliferation and induces apoptosis in bladder cancer. Oncotarget 2015, 6, 1064–1078. [Google Scholar] [CrossRef]
- Lu, M.; Ge, Q.; Wang, G.; Luo, Y.; Wang, X.; Jiang, W.; Liu, X.; Wu, C.L.; Xiao, Y.; Wang, X. CIRBP is a novel oncogene in human bladder cancer inducing expression of HIF-1alpha. Cell Death Dis. 2018, 9, 1046. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, G.; Peng, J.; Qian, G.; Jiang, W.; Xie, C.; Xiao, Y.; Wang, X. Knockdown of SIRT1 Suppresses Bladder Cancer Cell Proliferation and Migration and Induces Cell Cycle Arrest and Antioxidant Response through FOXO3a-Mediated Pathways. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhong, G.; Jiang, N.; Wang, B.; Fan, X.; Chen, C.; Chen, X.; Huang, J.; Lin, T. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J. Clin. Investig. 2018, 128, 861–875. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG database. In InNovartis Foundation Symposium; John Wiley: New York, NY, USA, 1999; pp. discussion 101–103, 119–128, 244–152. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Blaveri, E.; Simko, J.P.; Korkola, J.E.; Brewer, J.L.; Baehner, F.; Mehta, K.; Devries, S.; Koppie, T.; Pejavar, S.; Carroll, P.; et al. Bladder cancer outcome and subtype classification by gene expression. Clin. Cancer Res. 2005, 11, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
- Dyrskjot, L.; Kruhoffer, M.; Thykjaer, T.; Marcussen, N.; Jensen, J.L.; Moller, K.; Orntoft, T.F. Gene expression in the urinary bladder: A common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004, 64, 4040–4048. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Leem, S.H.; Lee, S.Y.; Kim, S.C.; Park, E.S.; Kim, S.B.; Kim, S.K.; Kim, Y.J.; Kim, W.J.; Chu, I.S. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J. Clin. Oncol. 2010, 28, 2660–2667. [Google Scholar] [CrossRef]
- Modlich, O.; Prisack, H.B.; Pitschke, G.; Ramp, U.; Ackermann, R.; Bojar, H.; Vogeli, T.A.; Grimm, M.O. Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: Use of cDNA array analysis of gene expression profiles. Clin. Cancer Res. 2004, 10, 3410–3421. [Google Scholar] [CrossRef]
- Sanchez-Carbayo, M.; Socci, N.D.; Lozano, J.; Saint, F.; Cordon-Cardo, C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006, 24, 778–789. [Google Scholar] [CrossRef]
- Tao, S.; Meng, S.; Zheng, X.; Xie, L. ATM participates in the regulation of viability and cell cycle via ellipticine in bladder cancer. Mol. Med. Rep. 2017, 15, 1143–1148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Hainaut, P.; Soussi, T.; Shomer, B.; Hollstein, M.; Greenblatt, M.; Hovig, E.; Harris, C.C.; Montesano, R. Database of p53 gene somatic mutations in human tumors and cell lines: Updated compilation and future prospects. Nucleic Acids Res. 1997, 25, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Zhang, Q.; Zhang, M.; Han, X.; Li, Q.; Xie, T.; Wu, Q.; Sui, X. p53/PCDH17/Beclin-1 Proteins as Prognostic Predictors for Urinary Bladder Cancer. J. Cancer 2019, 10, 6207–6216. [Google Scholar] [CrossRef] [PubMed]
- Villani, S.; Gagliano, N.; Procacci, P.; Sartori, P.; Comar, M.; Provenzano, M.; Favi, E.; Ferraresso, M.; Ferrante, P.; Delbue, S. Characterization of an in vitro model to study the possible role of polyomavirus BK in prostate cancer. J. Cell. Physiol. 2019, 234, 11912–11922. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Mei, L.; Fan, X.; Tang, C.; Ji, X.; Hu, X.; Shi, W.; Qian, Y.; Hussain, M.; Wu, J.; et al. Phosphodiesterase 5/protein kinase G signal governs stemness of prostate cancer stem cells through Hippo pathway. Cancer Lett. 2016, 378, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, R.; Arora, H.; Rizvi, M.A. EMT in cervical cancer: Its role in tumour progression and response to therapy. Cancer Lett. 2015, 356, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Erben, P.; Wezel, F.; Wirtz, R.; Martini, T.; Stein, D.; Weis, C.A.; Hartmann, A.; Bolenz, C. Role of the human ErbB family receptors in urothelial carcinoma of the bladder: MRNA expression status and prognostic relevance. Aktuelle Urol. 2017, 48, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Cronise, K.E.; Hernandez, B.G.; Gustafson, D.L.; Duval, D.L. Identifying the ErbB/MAPK Signaling Cascade as a Therapeutic Target in Canine Bladder Cancer. Mol. Pharmacol. 2019, 96, 36–46. [Google Scholar] [CrossRef]
- Han, Y.; Jin, X.; Zhou, H.; Liu, B. Identification of key genes associated with bladder cancer using gene expression profiles. Oncol. Lett. 2018, 15, 297–303. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, L.; Song, Z.; Xiong, M.; Zhang, Y.; Yang, Y.; Chen, K.; Chen, Z. The identification of new biomarkers for bladder cancer: A study based on TCGA and GEO datasets. J. Cell. Physiol. 2019, 234, 15607–15618. [Google Scholar] [CrossRef]
- Jia, Z.; Ai, X.; Sun, F.; Zang, T.; Guan, Y.; Gao, F. Identification of new hub genes associated with bladder carcinoma via bioinformatics analysis. Tumori 2015, 101, 117–122. [Google Scholar] [CrossRef]
- Han, Y.; Zheng, Q.; Tian, Y.; Ji, Z.; Ye, H. Identification of a nine-gene panel as a prognostic indicator for recurrence with muscle-invasive bladder cancer. J. Surg. Oncol. 2019, 119, 1145–1154. [Google Scholar] [CrossRef]
- Jiang, W.D.; Yuan, P.C. Molecular network-based identification of competing endogenous RNAs in bladder cancer. PLoS ONE 2019, 14, e0220118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, L.; Zang, Y.; Xu, Z. Identification of Core Genes and Key Pathways via Integrated Analysis of Gene Expression and DNA Methylation Profiles in Bladder Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 3024–3033. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, Y.; Chen, M.; Wang, S.; Wen, X.; Zhang, S. Identification of key candidate genes and biological pathways in bladder cancer. PeerJ 2018, 6, e6036. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhao, X.; Zhou, J.; Cheng, X.; Ye, Z.; Ji, Z. Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol. Ther. 2018, 19, 1039–1056. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, K.; Sorensen, K.D.; Brems-Eskildsen, A.S.; Modin, C.; Gaustadnes, M.; Hein, A.M.; Kruhoffer, M.; Laurberg, S.; Borre, M.; Wang, K.; et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell. Proteom. MCP 2008, 7, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, R.; Gao, L.; Cui, L.; Zhao, Z.; Yu, X.; Yuan, Y.; Xu, X. CASQ2 variants in Chinese children with catecholaminergic polymorphic ventricular tachycardia. Mol. Genet. Genom. Med. 2019, 7, e949. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Wu, J.; Li, W.; Chen, Z.; Yang, L. Progression of the role of CRYAB in signaling pathways and cancers. OncoTargets Ther. 2019, 12, 4129–4139. [Google Scholar] [CrossRef]
- Tian, Z.; Cao, S.; Li, C.; Xu, M.; Wei, H.; Yang, H.; Sun, Q.; Ren, Q.; Zhang, L. LncRNA PVT1 regulates growth, migration, and invasion of bladder cancer by miR-31/ CDK1. J. Cell. Physiol. 2019, 234, 4799–4811. [Google Scholar] [CrossRef]
- Shi, Y.X.; Zhu, T.; Zou, T.; Zhuo, W.; Chen, Y.X.; Huang, M.S.; Zheng, W.; Wang, C.J.; Li, X.; Mao, X.Y.; et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget 2016, 7, 85235–85243. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, Y.; Lee, J.H.; Kim, Y.S. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. Int. J. Pathol. 2013, 463, 681–687. [Google Scholar] [CrossRef]
- Wu, M.; Liu, Z.; Zhang, A.; Li, N. Identification of key genes and pathways in hepatocellular carcinoma: A preliminary bioinformatics analysis. Medicine 2019, 98, e14287. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, X.; Li, J.; He, Y. MicroRNA-144 inhibits cell proliferation, migration and invasion in human hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int. 2019, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Zeng, S.; Lu, X.; Xiong, Q.; Xue, Y.; Tong, L.; Xu, W.; Sun, Y.; Zhang, Z.; Xu, C. Overexpression of G2 and S phase-expressed-1 contributes to cell proliferation, migration, and invasion via regulating p53/FoxM1/CCNB1 pathway and predicts poor prognosis in bladder cancer. Int. J. Biol. Macromol. 2019, 123, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Roh, Y.G.; Park, K.; Kang, T.H.; Kim, W.J.; Lee, J.S.; Leem, S.H.; Chu, I.S. Expression signature defined by FOXM1-CCNB1 activation predicts disease recurrence in non-muscle-invasive bladder cancer. Clin. Cancer Res. 2014, 20, 3233–3243. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, P.; Wen, W.; Grubbs, C.J.; Townsend, R.R.; Malone, J.P.; Lubet, R.A.; You, M. Cross-species comparison of orthologous gene expression in human bladder cancer and carcinogen-induced rodent models. Am. J. Transl. Res. 2010, 3, 8–27. [Google Scholar] [PubMed]
- Li, J.; Ying, Y.; Xie, H.; Jin, K.; Yan, H.; Wang, S.; Xu, M.; Xu, X.; Wang, X.; Yang, K.; et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. FASEB J. 2019, 33, 1374–1388. [Google Scholar] [CrossRef]
- Bai, Y.; Xiong, L.; Zhu, M.; Yang, Z.; Zhao, J.; Tang, H. Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark. Sect. A Dis. Markers 2019, 24, 371–382. [Google Scholar] [CrossRef]
- Gan, H.; Lin, L.; Hu, N.; Yang, Y.; Gao, Y.; Pei, Y.; Chen, K.; Sun, B. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem. Funct. 2019, 37, 424–431. [Google Scholar] [CrossRef]
- Imai, T.; Oue, N.; Nishioka, M.; Mukai, S.; Oshima, T.; Sakamoto, N.; Sentani, K.; Matsusaki, K.; Yoshida, K.; Yasui, W. Overexpression of KIF11 in Gastric Cancer with Intestinal Mucin Phenotype. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2017, 84, 16–24. [Google Scholar] [CrossRef]
- Jiang, M.; Zhuang, H.; Xia, R.; Gan, L.; Wu, Y.; Ma, J.; Sun, Y.; Zhuang, Z. KIF11 is required for proliferation and self-renewal of docetaxel resistant triple negative breast cancer cells. Oncotarget 2017, 8, 92106–92118. [Google Scholar] [CrossRef]
- Pan, S.; Zhan, Y.; Chen, X.; Wu, B.; Liu, B. Identification of Biomarkers for Controlling Cancer Stem Cell Characteristics in Bladder Cancer by Network Analysis of Transcriptome Data Stemness Indices. Front. Oncol. 2019, 9, 613. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yang, L.; Zhang, Z.; Yu, J.; Dai, L.; Gao, M.; Shang, Z.; Niu, Y. KIF20A Affects the Prognosis of Bladder Cancer by Promoting the Proliferation and Metastasis of Bladder Cancer Cells. Dis. Markers 2019, 2019, 4863182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, P.; Liu, J.; Broaddus, R.R.; Xue, F.; Zhang, W. Centrosome-associated regulators of the G(2)/M checkpoint as targets for cancer therapy. Mol. Cancer 2009, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, T.; Tanikawa, C.; Furukawa, Y.; Katagiri, T.; Nakamura, Y.; Matsuda, K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008, 27, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Gayyed, M.F.; El-Maqsoud, N.M.; Tawfiek, E.R.; El Gelany, S.A.; Rahman, M.F. A comprehensive analysis of CDC20 overexpression in common malignant tumors from multiple organs: Its correlation with tumor grade and stage. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 749–762. [Google Scholar] [CrossRef]
- Morikawa, T.; Kawai, T.; Abe, H.; Kume, H.; Homma, Y.; Fukayama, M. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int. J. Clin. Exp. Pathol. 2013, 6, 1367–1374. [Google Scholar]
- Kim, W.T.; Jeong, P.; Yan, C.; Kim, Y.H.; Lee, I.S.; Kang, H.W.; Kim, Y.J.; Lee, S.C.; Kim, S.J.; Kim, Y.T.; et al. UBE2C cell-free RNA in urine can discriminate between bladder cancer and hematuria. Oncotarget 2016, 7, 58193–58202. [Google Scholar] [CrossRef]
- Jin, S.; Wang, X.; Tong, T.; Zhang, D.; Shi, J.; Chen, J.; Zhan, Q. Aurora-A enhances malignant development of esophageal squamous cell carcinoma (ESCC) by phosphorylating beta-catenin. Mol. Oncol. 2015, 9, 249–259. [Google Scholar] [CrossRef]
- Chen, C.; Song, G.; Xiang, J.; Zhang, H.; Zhao, S.; Zhan, Y. AURKA promotes cancer metastasis by regulating epithelial-mesenchymal transition and cancer stem cell properties in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 486, 514–520. [Google Scholar] [CrossRef]
- Guo, M.; Lu, S.; Huang, H.; Wang, Y.; Yang, M.Q.; Yang, Y.; Fan, Z.; Jiang, B.; Deng, Y. Increased AURKA promotes cell proliferation and predicts poor prognosis in bladder cancer. BMC Syst. Biol. 2018, 12, 118. [Google Scholar] [CrossRef]
- Yuan, Q.; Sun, T.; Ye, F.; Kong, W.; Jin, H. MicroRNA-124-3p affects proliferation, migration and apoptosis of bladder cancer cells through targeting AURKA. Cancer Biomark. Sect. A Dis. Markers 2017, 19, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Su, B.; Fang, D.; Tang, Y.; Xiong, G.; Guo, Z.; He, Q.; Yang, X.; Zhao, W.; Guo, Y.; et al. High expression of KPNA2 defines poor prognosis in patients with upper tract urothelial carcinoma treated with radical nephroureterectomy. BMC Cancer 2015, 15, 380. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, D.; Cheng, R.; Wang, Y.; Jiang, S.; Zhu, Y.; Fan, L.; Mao, X.; Gui, Y.; Li, Z.; et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget 2016, 7, 72767–72776. [Google Scholar] [CrossRef] [PubMed][Green Version]
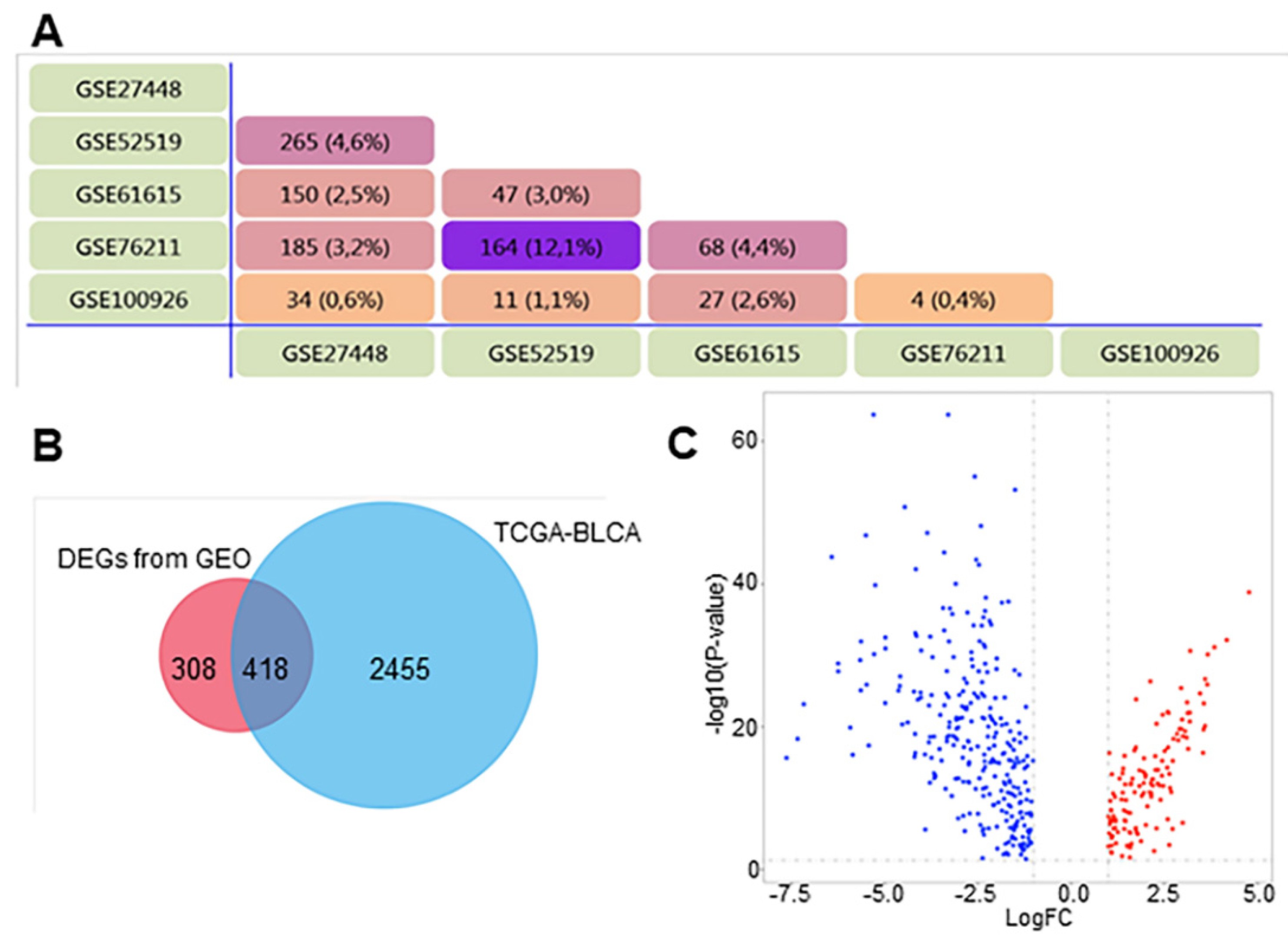

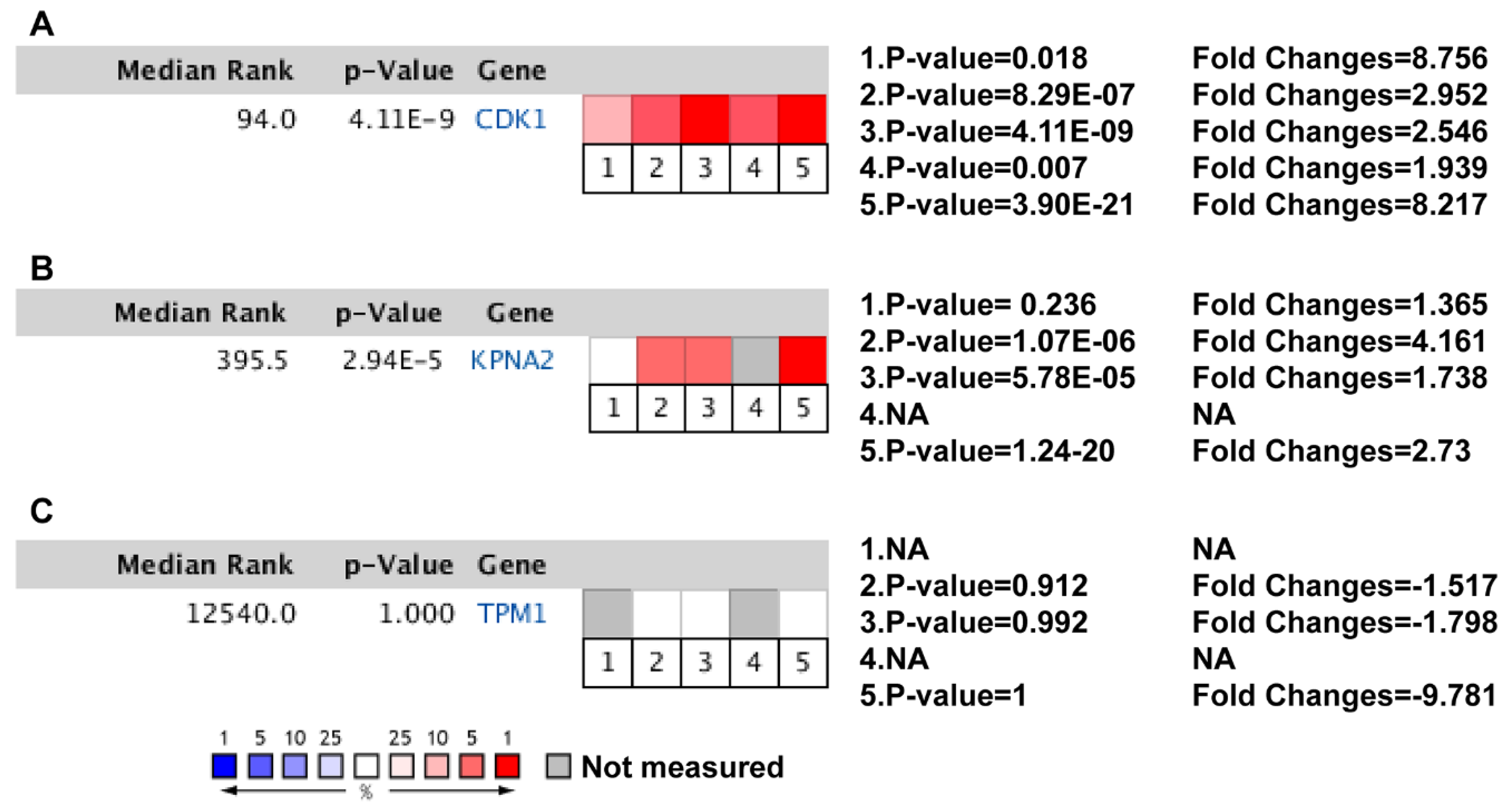
| Dataset | Number of Noncancerous Bladder Tissue Samples | Number of Cancer Tissue Samples | Number of DEGs | Platform | Manufacturer of Platform |
|---|---|---|---|---|---|
| GSE27448 | 5 | 10 | 4701 | GPL2895 | GE Healthcare |
| GSE52519 | 3 | 9 | 742 | GPL16884 | JPT Peptide Technology |
| GSE61615 | 2 | 2 | 736 | GPL14550 | Agilent Technologies |
| GSE76211 | 3 | 3 | 658 | GPL17586 | Affymetrix |
| GSE100926 | 3 | 3 | 194 | GPL16956 | Agilent Technologies |
| Gene Symbol | Other Names | Full Name | Role of Genes | Putative or Observed Effect |
|---|---|---|---|---|
| CDK1 | CDC2, CDC28A, P34CDC2 | Cyclin-dependent kinase 1 | Oncogene | Overexpression in BCa; Activating CDK1 facilitates the proliferation and invasion of BCa. [23] |
| CCNB1 | cyclin B1 | CCNB | Oncogene | Increased expression in BCa; Regulating BCa through the FoxM1/CCNB1 pathway, correlates with poor prognosis. [24] |
| KIF11 | kinesin family member 11 | EG5; HKSP; KNSL1; MCLMR; TRIP5 | Oncogene | Overexpression in BCa; Predicted to be Oncogene. [25] |
| CCNA2 | cyclin A2 | CCN1; CCNA | Oncogene | Over expression in BCa; Activity of CCNA2 regulated the EMT via ROCK/AKT/β-catenin/SNAIL pathway to influence the prognosis of BCa. [26] |
| UBE2C | ubiquitin conjugating enzyme E2 C | UBCH10; dJ447F3.2 | Oncogene | High expression in BCa; Activating UBE2C induced progression and correlates with poor survival. [27,28] |
| CDC20 | cell division cycle 20 | CDC20A; p55CDC; bA276H19.3 | Oncogene | Increased expression in BCa; Activating CDC20 increased carcinogenesis and correlates with poor survival in BCa. [29] |
| MAD2L1 | mitotic arrest deficient 2 like 1 | MAD2; HSMAD2 | Oncogene | Overexpression in BCa; Activating MAD2 was associated with incidence of recurrence and progression of BCa. [30] |
| KIF2C | kinesin family member 2C | MCAK; CT139; KNSL6 | Oncogene | Overexpression of KIF2C was confirmed in BCa from rat model. [31] |
| AURKA | aurora kinase A | AIK; ARK1; AURA; BTAK; STK6; STK7; STK15; PPP1R47 | Oncogene | Highly expression in BCa; Activating AURKA is associated with poor prognosis, tumor stage and grade. [32,33] |
| KIF20A | kinesin family member 20A | MKLP2; RAB6KIFL | Oncogene | Over expression in BCa; KIF20A was associated with the development of BCa and correlated with poor survival. [34] |
| KPNA2 | karyopherin subunit alpha 2 | QIP2; RCH1; IPOA1; SRP1alpha; SRP1-alpha | Oncogene | Increased expression in BCa; Activation of KPNA2 defines poor prognosis in BCa. [35,36] |
| TPM1 | tropomyosin 1 | CMH3; TMSA; CMD1Y; LVNC9; C15orf13; | Tumor suppressor | Downregulated in BCa; Activating TPM1 inhibited the proliferation and promoted the apoptosis of Bladder tumor cells. [37,38] |
| CASQ2 | calsequestrin 2 | PDIB2 | Disease-related gene | Reduced in BCa in present study; Predicted to be tumor suppressor; No study of CASQ2 reported in BCa. [39] |
| CRYAB | crystallin alpha B | MFM2; CRYA2; CTPP2; HSPB5; CMD1II; CTRCT16; | Cancer-related gene | Decreased expression in BCa in present study; Enforced activation of CRYAB correlated to poor prognosis of lung cancer and other tumors [40]; No study of CRYAB reported in BCa. |
| Hub Genes | Reference | ||
|---|---|---|---|
| Upregulated | Downregulated | Regulation not Specified | (Databases Used) |
| AURKA *, CCNA2 *, CCNE1#, CDC20 *, BCL3#, CEP55 *, DCUN1D1#, FGFR1OP#, HMMR *, MAP3K8#, MYB#, PTG1#, VEGFA# | DUSP26#, MEIS1 * | Han et al. [48] (GSE52519) | |
| CDC20 * | ACTA#, DCN#, MYH11 *, TPM1 *, VIM#, TGFB3# | Hu et al. [49] (GSE13507, TCGA-BLCA) | |
| ITGA7#, GRB14#, CDC20 *, PSMB1#, POLR2F/2H#, RPS14/15# | Jia et al. [50] (GSE27448) | ||
| EME1#, AKAP9#, ZNF91#, OARD3#, STAG2#, ZFP36L2#, METTL3#, POLR3#, MUC7# | Han et al. [51] (TCGA-BLCA) | ||
| DSN1#, KNTC1 *, CDK1 *, CENPM#, HIST1H2BJ#, EZH2 *, CENPF *, RAD51#, BRCA1#, EXO1# | Jiang and Yuan [52] (Circular RNA Interactome database; TCGA) | ||
| CDH1#, DDOST#, CASP8#, DHX15#, PTRF@ | GNG4#, ADCY9#, NPY#, ADRA2B#, PENK# | Zhang et al. [53] (GSE3167, GSE65635, GSE33510) | |
| TOP2A *, CDC20 *, UBE2C *, CCNB1 *, CCNB2 *, VEGFA#, AURKB *, AURKA *, CEP55 * | ACTA2# | Gao et al. [54] (GSE7476, GSE13507, GSE37815, GSE65635) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Berndt-Paetz, M.; Neuhaus, J. Identification of Key Biomarkers in Bladder Cancer: Evidence from a Bioinformatics Analysis. Diagnostics 2020, 10, 66. https://doi.org/10.3390/diagnostics10020066
Zhang C, Berndt-Paetz M, Neuhaus J. Identification of Key Biomarkers in Bladder Cancer: Evidence from a Bioinformatics Analysis. Diagnostics. 2020; 10(2):66. https://doi.org/10.3390/diagnostics10020066
Chicago/Turabian StyleZhang, Chuan, Mandy Berndt-Paetz, and Jochen Neuhaus. 2020. "Identification of Key Biomarkers in Bladder Cancer: Evidence from a Bioinformatics Analysis" Diagnostics 10, no. 2: 66. https://doi.org/10.3390/diagnostics10020066
APA StyleZhang, C., Berndt-Paetz, M., & Neuhaus, J. (2020). Identification of Key Biomarkers in Bladder Cancer: Evidence from a Bioinformatics Analysis. Diagnostics, 10(2), 66. https://doi.org/10.3390/diagnostics10020066