Noninvasive Measurement of [11C]PiB Distribution Volume Using Integrated PET/MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. PET/MRI Scanner
2.3. PET and MRI Image Acquisition and PET Reconstruction
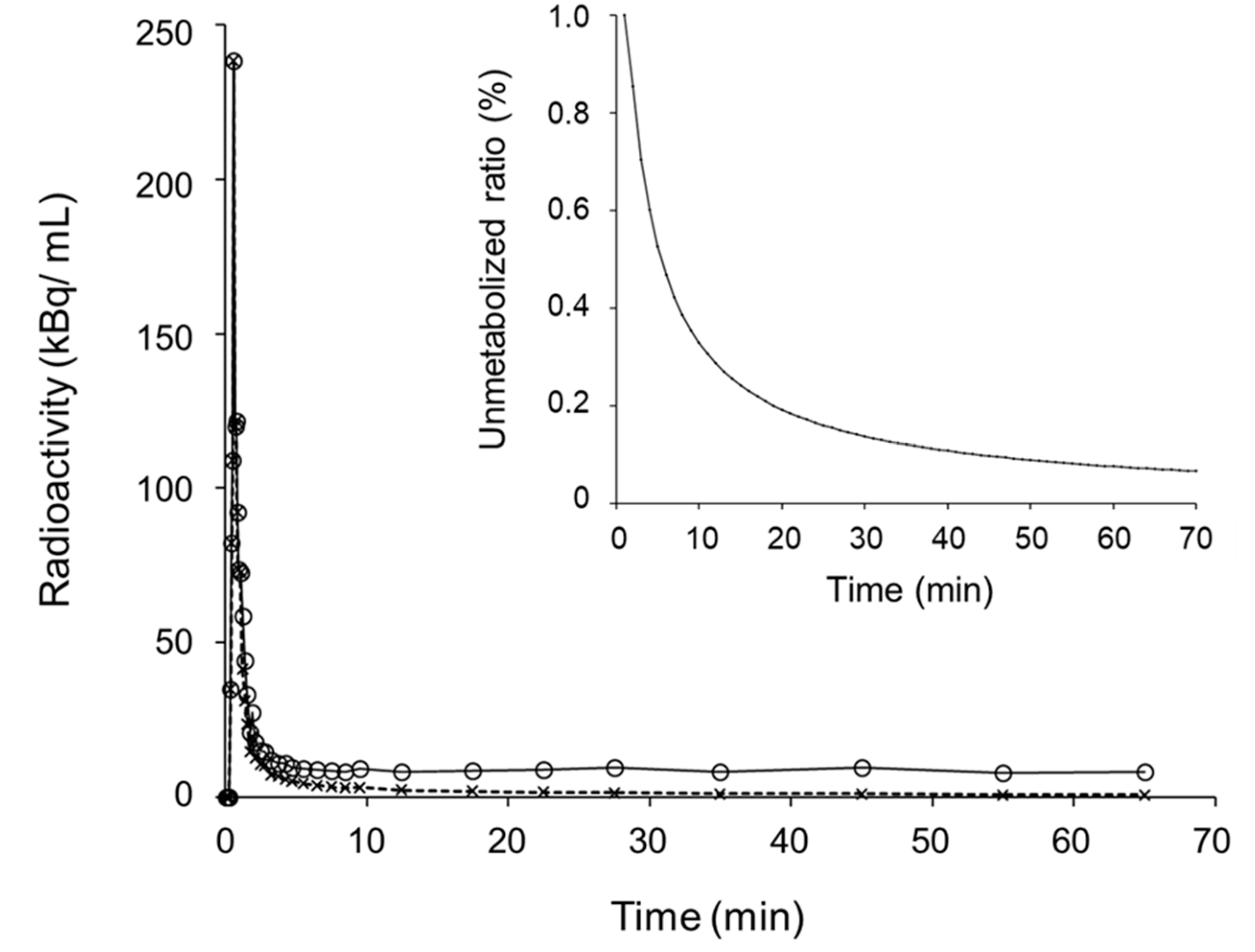
2.4. Distribution Volume and SUV Image Calculation
2.5. Statistical Analyses
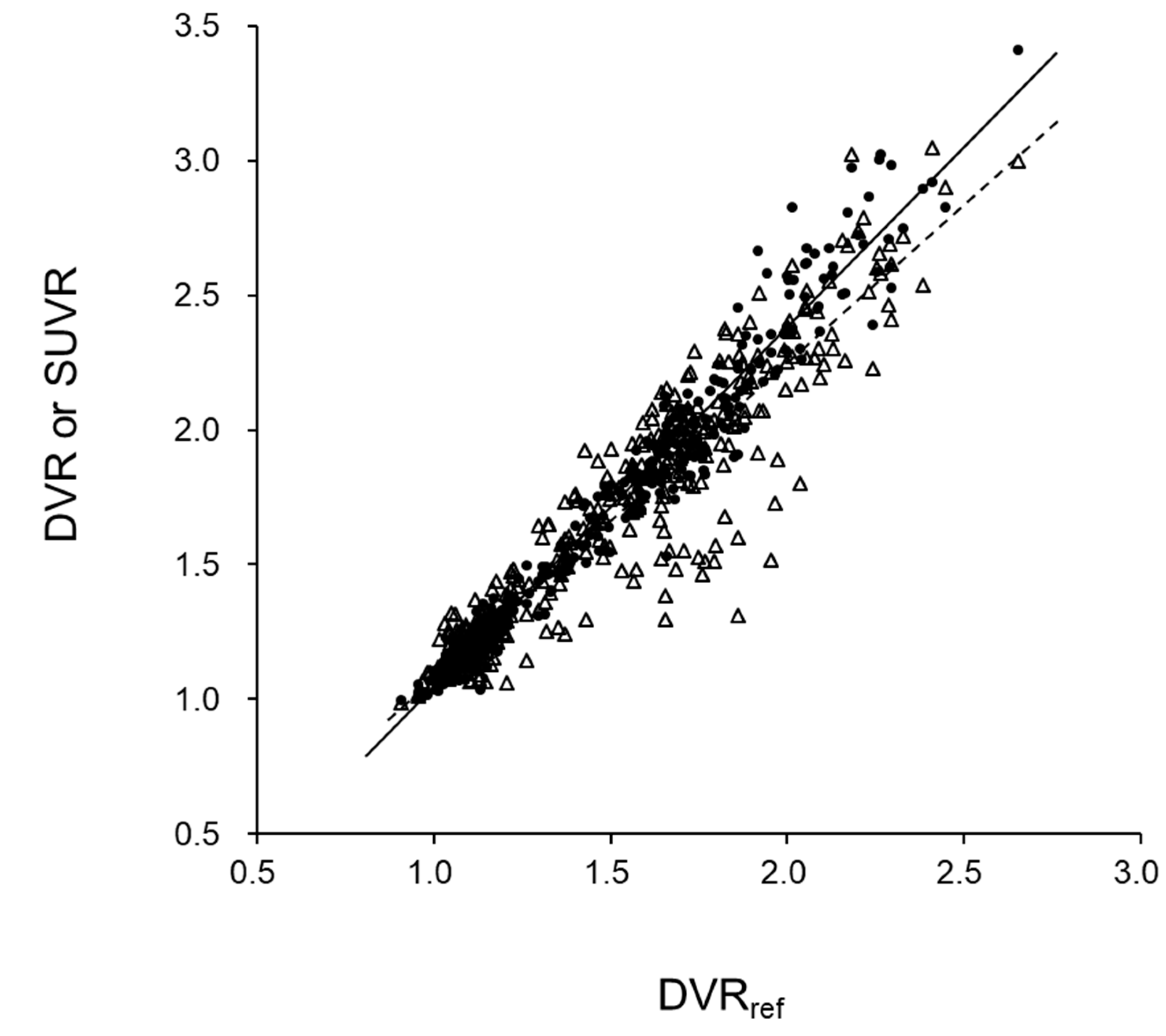
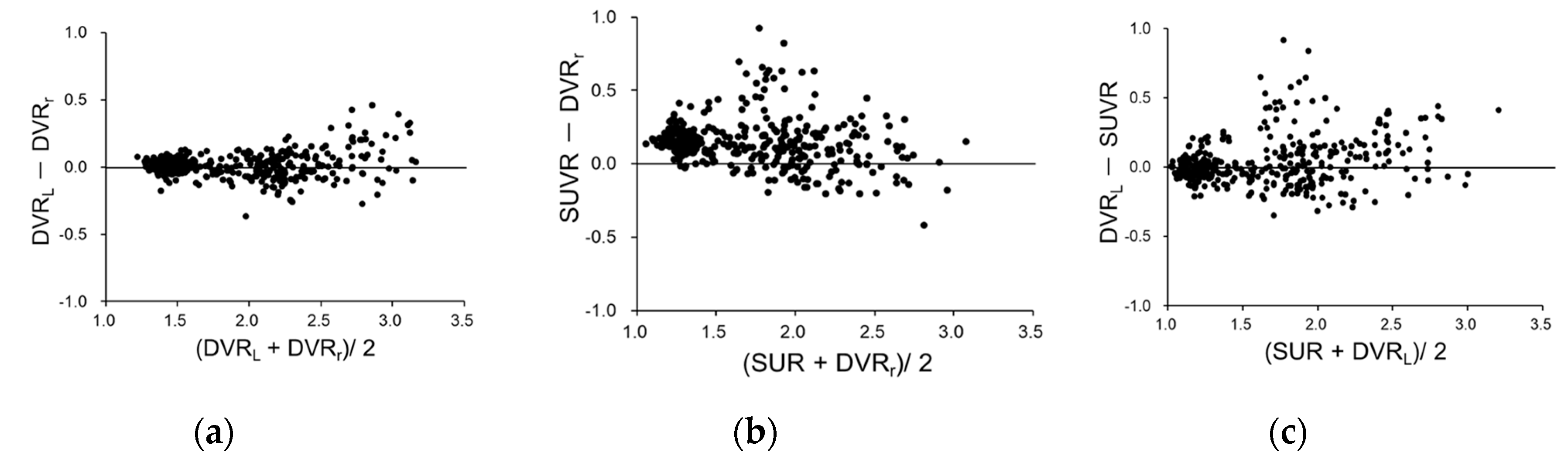
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Lopresti, B.J.; Klunk, W.E.; Mathis, C.A.; Hoge, J.A.; Ziolko, S.K.; Lu, X.; Meltzer, C.C.; Schimmel, K.; Tsopelas, N.D.; DeKosky, S.T.; et al. Simplified quantification of Pittsburgh Compound B amyloid imaging PET studies: A comparative analysis. J. Nucl. Med. 2005, 46, 1959–1972. Available online: http://jnm.snmjournals.org/content/46/12/1959.full.pdf+html (accessed on 24 October 2020).
- Yaqub, M.; Tolboom, N.; Boellaard, R.; van Berckel, B.N.; van Tilburg, E.W.; Luurtsema, G.; Scheltens, P.; Lammertsma, A.A. Simplified parametric methods for [11C]PIB studies. Neuroimage 2008, 42, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Price, J.C.; Klunk, W.E.; Lopresti, B.J.; Lu, X.; Hoge, J.A.; Ziolko, S.K.; Holt, D.P.; Meltzer, C.C.; DeKosky, S.T.; Mathis, C.A. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J. Cereb. Blood Flow Metab. 2005, 25, 1528–1547. [Google Scholar] [CrossRef] [PubMed]
- Mourik, J.E.; Lubberink, M.; Schuitemaker, A.; Tolboom, N.; van Berckel, B.N.; Lammertsma, A.A.; Boellaard, R. Image-derived input functions for PET brain studies. Eur. J. Nucl. Med. Mol. Imaging 2009, 36, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Okazawa, H.; Higashino, Y.; Tsujikawa, T.; Arishima, H.; Mori, T.; Kiyono, Y.; Kimura, H.; Kikuta, K. Noninvasive method for measurement of cerebral blood flow using O-15 water PET/MRI with ASL correlation. Eur. J. Radiol. 2018, 105, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Okazawa, H.; Tsujikawa, T.; Higashino, Y.; Kikuta, K.; Mori, T.; Makino, A.; Kiyono, Y. No significant difference found in PET/MRI CBF values reconstructed with CT-atlas-based and ZTE MR attenuation correction. Eur. J. Nucl. Med. Mol. Imaging Res. 2019, 9, 26. [Google Scholar] [CrossRef]
- Gunn, R.N.; Sargent, P.A.; Bench, C.J.; Rabiner, E.A.; Osman, S.; Pike, V.W.; Hume, S.P.; Grasby, P.M.; Lammertsma, A.A. Tracer kinetic modeling of the 5-HT1A receptor ligand [carbonyl-11C]WAY-100635 for PET. Neuroimage 1998, 8, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, C.; Ishii, K.; Kimura, Y.; Hyodo, T.; Hosono, M.; Sakaguchi, K.; Usami, K.; Shimamoto, K.; Yamazoe, Y.; Murakami, T. Performance of 11C-Pittsburgh Compound B PET Binding Potential Images in the Detection of Amyloid Deposits on Equivocal Static Images. J. Nucl. Med. 2015, 56, 1910–1915. [Google Scholar] [PubMed]
- Yamane, T.; Ishii, K.; Sakata, M.; Ikari, Y.; Nishio, T.; Ishii, K.; Kato, T.; Ito, K.; Senda, M.; J-ADNI Study Group. Inter-rater variability of visual interpretation and comparison with quantitative evaluation of 11C-PiB PET amyloid images of the Japanese Alzheimer’s Disease Neuroimaging Initiative (J-ADNI) multicenter study. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 850–857. [Google Scholar] [PubMed]
- Levin, C.S.; Maramraju, S.H.; Khalighi, M.M.; Deller, T.W.; Delso, G.; Jansen, F. Design features and mutual compatibility studies of the time-of-flight PET capable GE SIGNA PET/MR system. IEEE Trans. Med. Imaging 2016, 35, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Wiesinger, F.; Sacolick, L.I.; Menini, A.; Kaushik, S.S.; Ahn, S.; Veit-Haibach, P.; Delso, G.; Shanbhag, D.D. Zero TE MR bone imaging in the head. Magn. Reson. Med. 2016, 75, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Delso, G.; Kemp, B.; Kaushik, S.; Wiesinger, F.; Sekine, T. Improving PET/MR brain quantitation with template-enhanced ZTE. Neuroimage 2018, 181, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Okazawa, H.; Ikawa, M.; Jung, M.; Maruyama, R.; Tsujikawa, T.; Mori, T.; Rahman, M.G.M.; Makino, A.; Kiyono, Y.; Kosaka, H. Multimodal analysis using [11C]PiB-PET/MRI for functional evaluation of patients with Alzheimer’s disease. EJNMMI Res. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Tsujikawa, T.; Mori, T.; Kiyono, Y.; Okazawa, H. Estimation of arterial input by a noninvasive image derived method in brain H215O PET study: Confirmation of arterial location using MR angiography. Phys. Med. Biol. 2017, 62, 4514–4524. [Google Scholar] [CrossRef] [PubMed]
- Logan, J.; Fowler, J.S.; Volkow, N.D.; Wolf, A.P.; Dewey, S.L.; Schlyer, D.J.; MacGregor, R.R.; Hitzemann, R.; Bendriem, B.; Gatley, S.J.; et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J. Cereb. Blood Flow Metab. 1990, 10, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Okazawa, H.; Yamauchi, H.; Sugimoto, K.; Magata, Y.; Kudo, T.; Yonekura, Y. Effects of metabolite correction for arterial input function on quantitative receptor images with 11C-flumazenil in clinical PET studies. J. Comput. Assist. Tomogr. 2004, 28, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Logan, J.; Fowler, J.S.; Volkow, N.D.; Wang, G.J.; Ding, Y.S.; Alexoff, D.L. Distribution volume ratios without blood sampling from graphical analysis of PET data. J. Cereb. Blood Flow Metab. 1996, 16, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.; Giuliani, A.; Öberg, J.; Chincarini, A.; Morbelli, S.; Brugnolo, A.; Arnaldi, D.; Picco, A.; Bauckneht, M.; Buschiazzo, A.; et al. Predicting the transition from normal aging to Alzheimer’s disease: A statistical mechanistic evaluation of FDG-PET data. Neuroimage 2016, 141, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Brugnolo, A.; De Carli, F.; Pagani, M.; Morbelli, S.; Jonsson, C.; Chincarini, A.; Frisoni, G.B.; Galluzzi, S.; Perneczky, R.; Drzezga, A.; et al. Head-to-head comparison among semi-quantification tools of brain FDG-PET to aid the diagnosis of prodromal Alzheimer’s disease. J. Alzheimers Dis. 2019, 68, 383–394. [Google Scholar] [CrossRef]


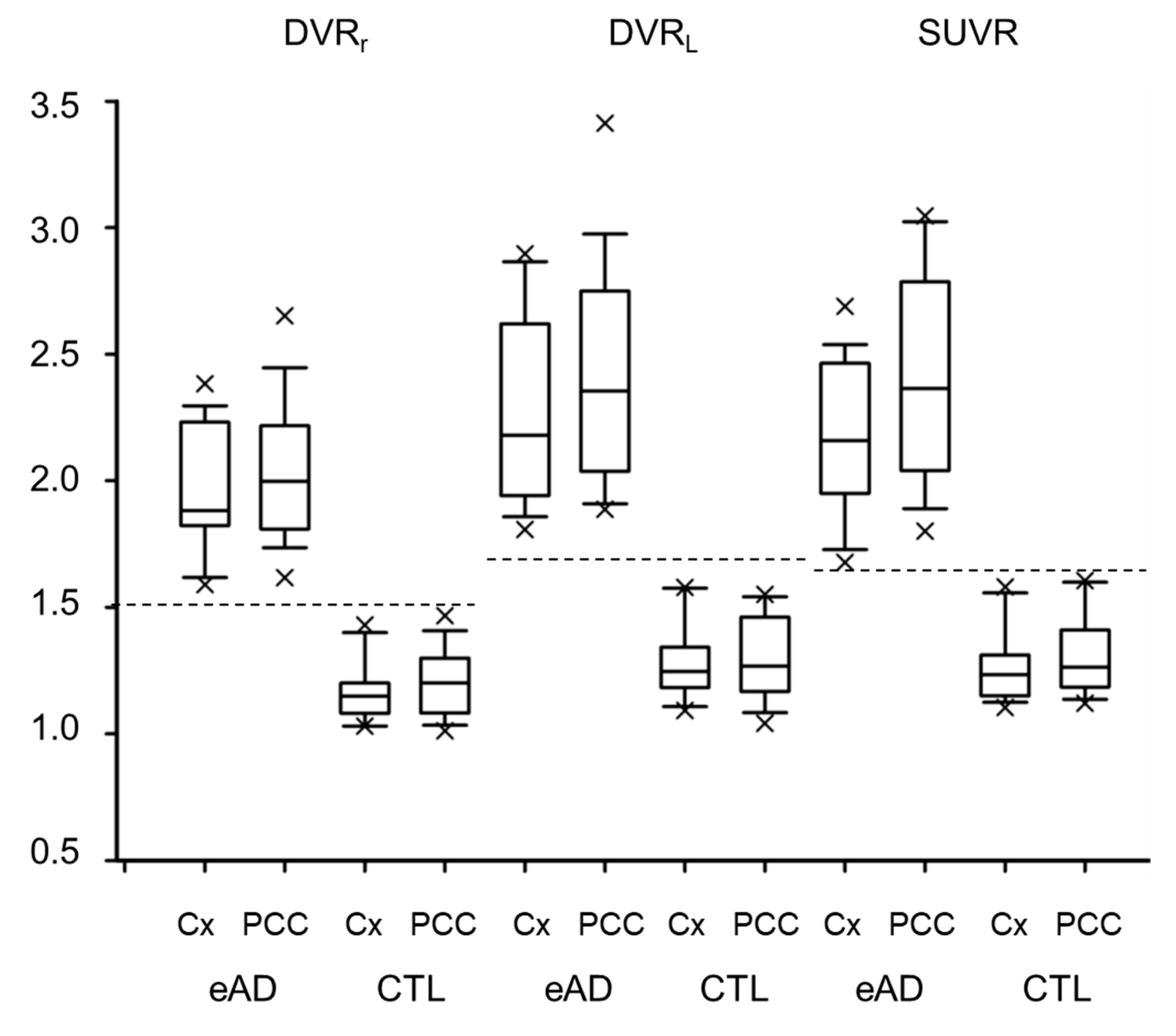
| DVRr | DVRL | SUVR | |||||
|---|---|---|---|---|---|---|---|
| CTL | eAD | CTL | eAD | CTL | eAD | ||
| Frontal | L | 1.10 ± 0.11 | 1.78 ± 0.23 †, * | 1.22 ± 0.14 | 2.14 ± 0.32 †, * | 1.20 ± 0.13 | 2.00 ± 0.30 †, * |
| R | 1.13 ± 0.10 | 1.79 ± 0.25 †, * | 1.24 ± 0.13 | 2.17 ± 0.38 † | 1.23 ± 0.13 | 2.04 ± 0.32 †, * | |
| Temporal | L | 1.07 ± 0.05 | 1.52 ± 0.16 †, * | 1.13 ± 0.06 | 1.72 ± 0.22 †, * | 1.17 ± 0.08 | 1.78 ± 0.27 †, * |
| R | 1.07 ± 0.05 | 1.53 ± 0.16 †, * | 1.13 ± 0.06 | 1.74 ± 0.23 †, * | 1.16 ± 0.08 | 1.79 ± 0.28 †, * | |
| Parietal | L | 1.11 ± 0.11 | 1.87 ± 0.22†,* | 1.18 ± 0.12 | 2.11 ± 0.31†,* | 1.18 ± 0.12 | 2.06 ± 0.32†,* |
| R | 1.13 ± 0.11 | 1.91 ± 0.27 † | 1.19 ± 0.13 | 2.19 ± 0.38 † | 1.21 ± 0.14 | 2.10 ± 0.32 †, * | |
| Occipital | L | 1.10 ± 0.05 | 1.57 ± 0.19 †, * | 1.16 ± 0.07 | 1.73 ± 0.25 †, * | 1.16 ± 0.06 | 1.73 ± 0.28 †, * |
| R | 1.11 ± 0.06 | 1.60 ± 0.17 †, * | 1.16 ± 0.08 | 1.78 ± 0.24 †, * | 1.19 ± 0.08 | 1.76 ± 0.21 †, * | |
| PCC | 1.20 ± 0.13 | 2.04 ± 0.28 † | 1.30 ± 0.16 | 2.43 ± 0.42 † | 1.31 ± 0.16 | 2.42 ± 0.41 † | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okazawa, H.; Ikawa, M.; Tsujikawa, T.; Makino, A.; Mori, T.; Kiyono, Y.; Kosaka, H. Noninvasive Measurement of [11C]PiB Distribution Volume Using Integrated PET/MRI. Diagnostics 2020, 10, 993. https://doi.org/10.3390/diagnostics10120993
Okazawa H, Ikawa M, Tsujikawa T, Makino A, Mori T, Kiyono Y, Kosaka H. Noninvasive Measurement of [11C]PiB Distribution Volume Using Integrated PET/MRI. Diagnostics. 2020; 10(12):993. https://doi.org/10.3390/diagnostics10120993
Chicago/Turabian StyleOkazawa, Hidehiko, Masamichi Ikawa, Tetsuya Tsujikawa, Akira Makino, Tetsuya Mori, Yasushi Kiyono, and Hirotaka Kosaka. 2020. "Noninvasive Measurement of [11C]PiB Distribution Volume Using Integrated PET/MRI" Diagnostics 10, no. 12: 993. https://doi.org/10.3390/diagnostics10120993
APA StyleOkazawa, H., Ikawa, M., Tsujikawa, T., Makino, A., Mori, T., Kiyono, Y., & Kosaka, H. (2020). Noninvasive Measurement of [11C]PiB Distribution Volume Using Integrated PET/MRI. Diagnostics, 10(12), 993. https://doi.org/10.3390/diagnostics10120993






