Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images
Abstract
1. Introduction
2. Materials and Methods
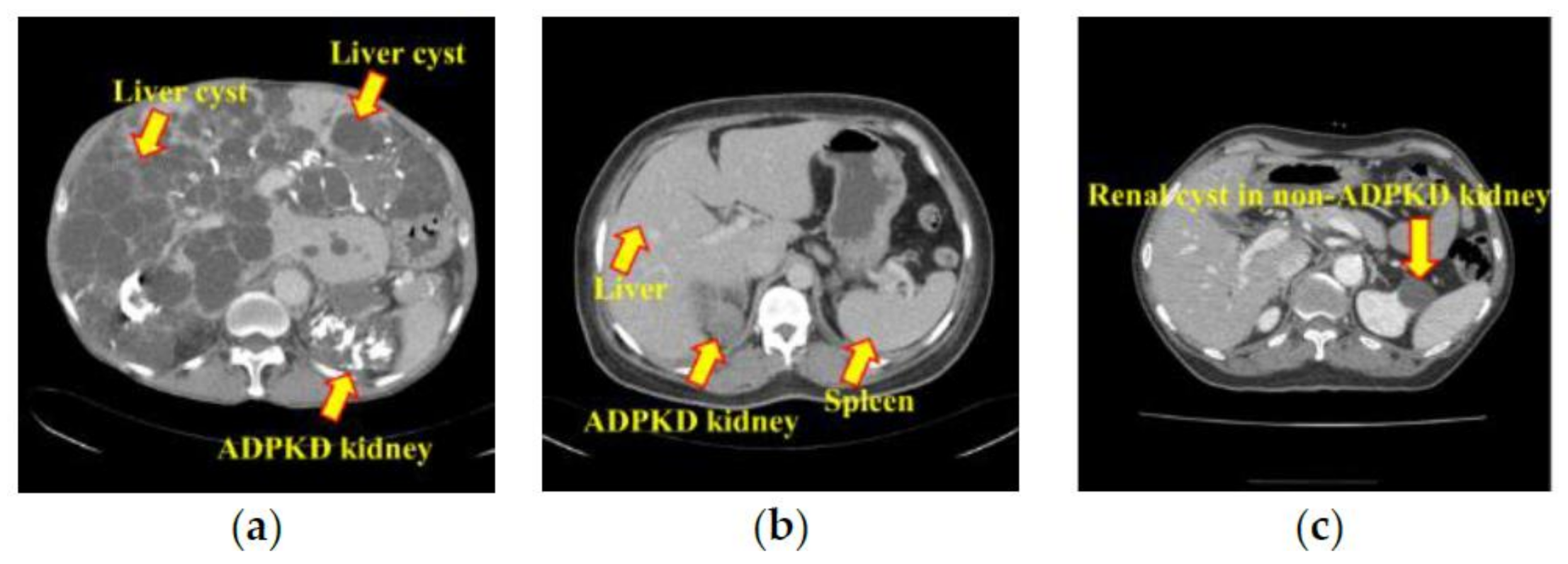
2.1. Data Acquisition
2.2. Ground Truth Annotation
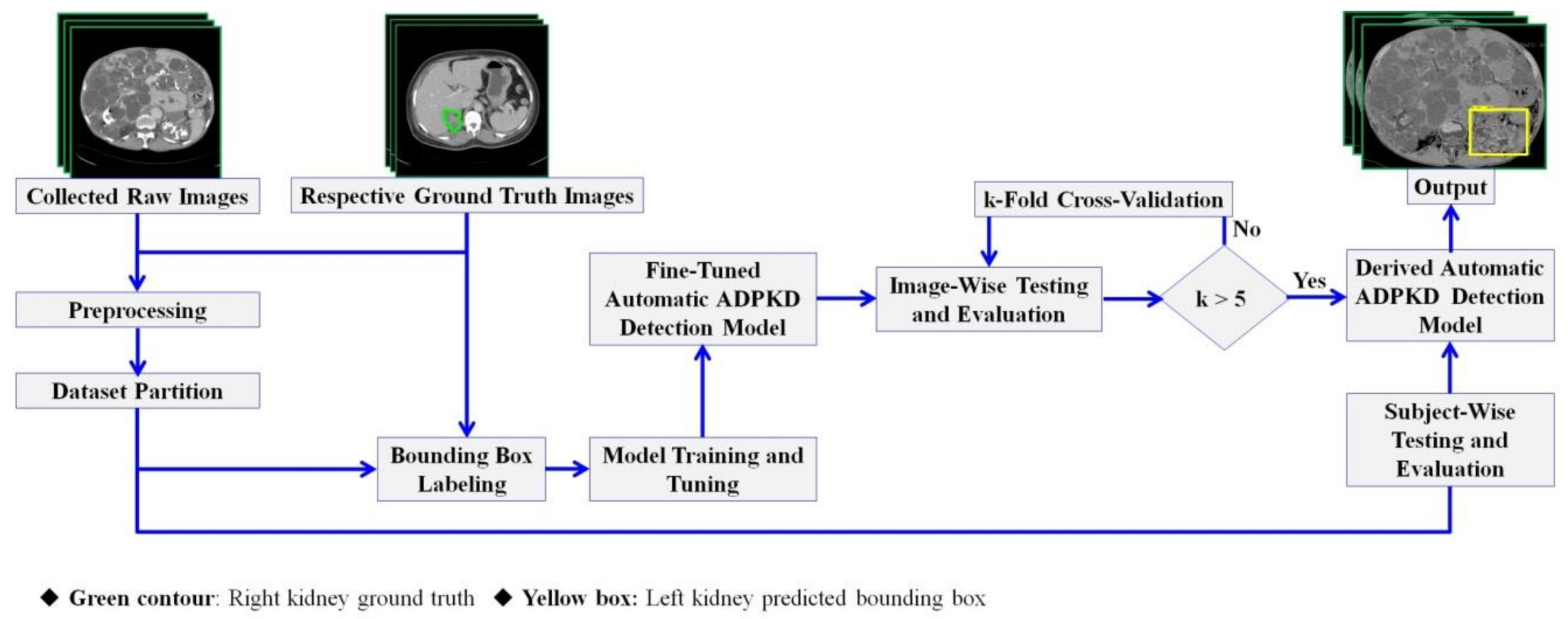
2.3. Methods
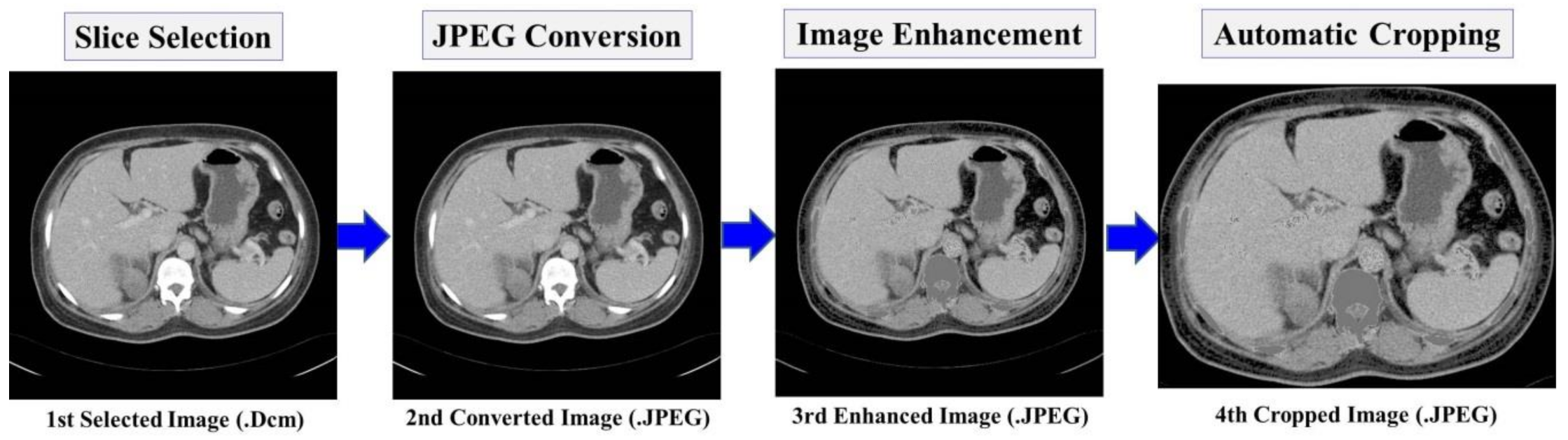
2.3.1. Preprocessing
2.3.2. Dataset Partition
2.3.3. Bounding Box Labeling
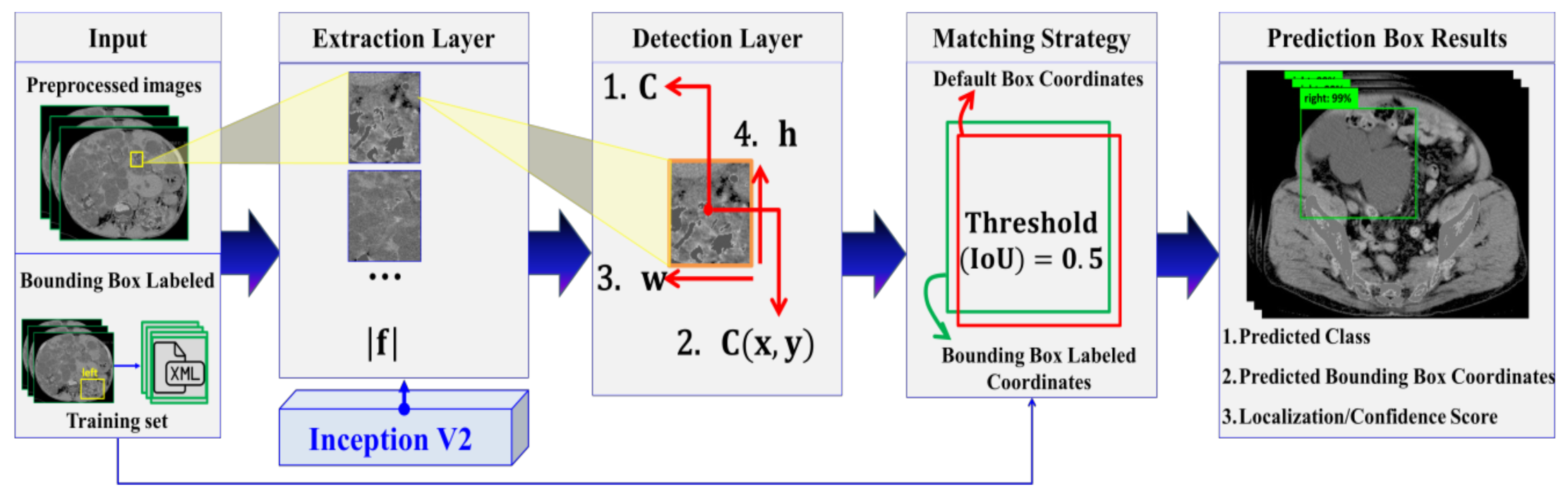
2.3.4. Automatic ADPKD Detection Model
2.3.5. Training and Tuning Model
2.3.6. Image-Wise and Subject-Wise Testing and Evaluation
2.4. Experimental Setup
2.5. Evaluation Metrics
2.6. Evaluation Procedures
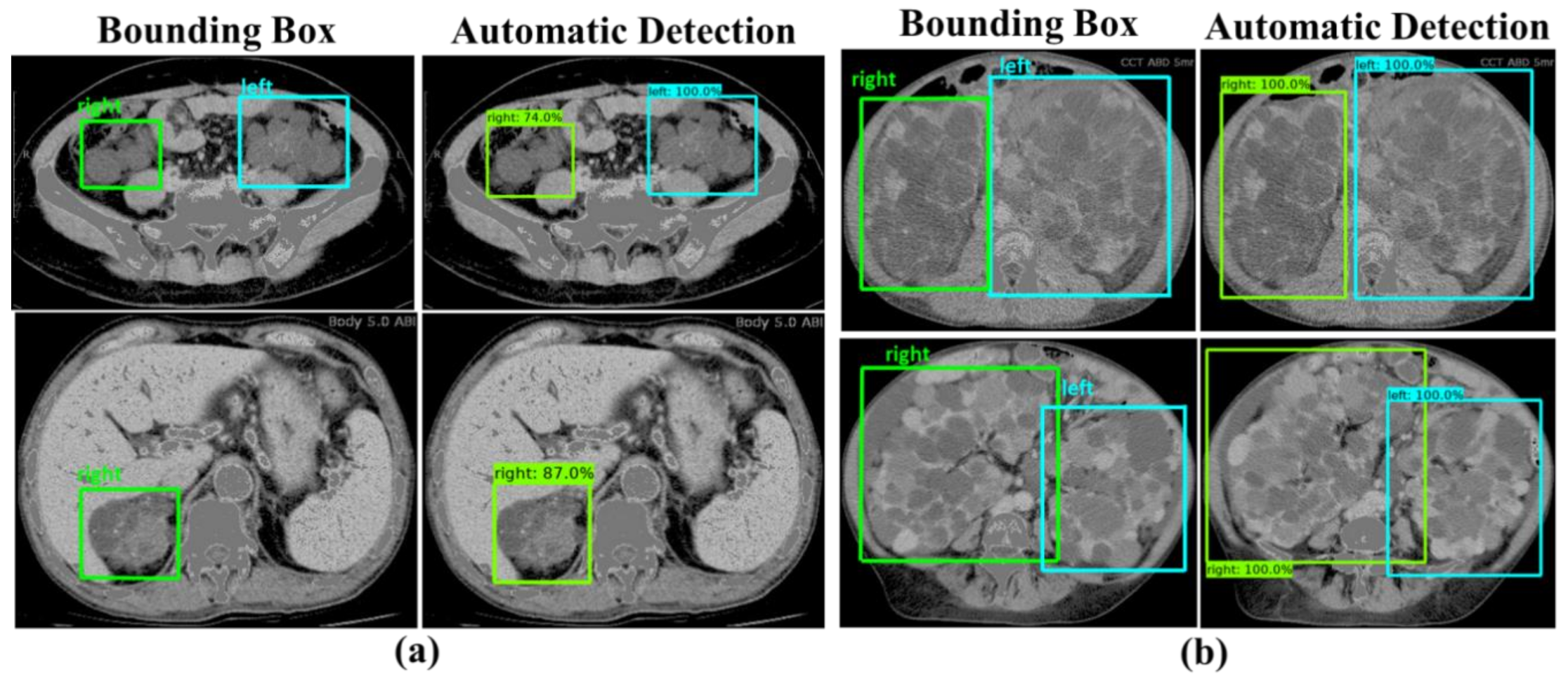
3. Results
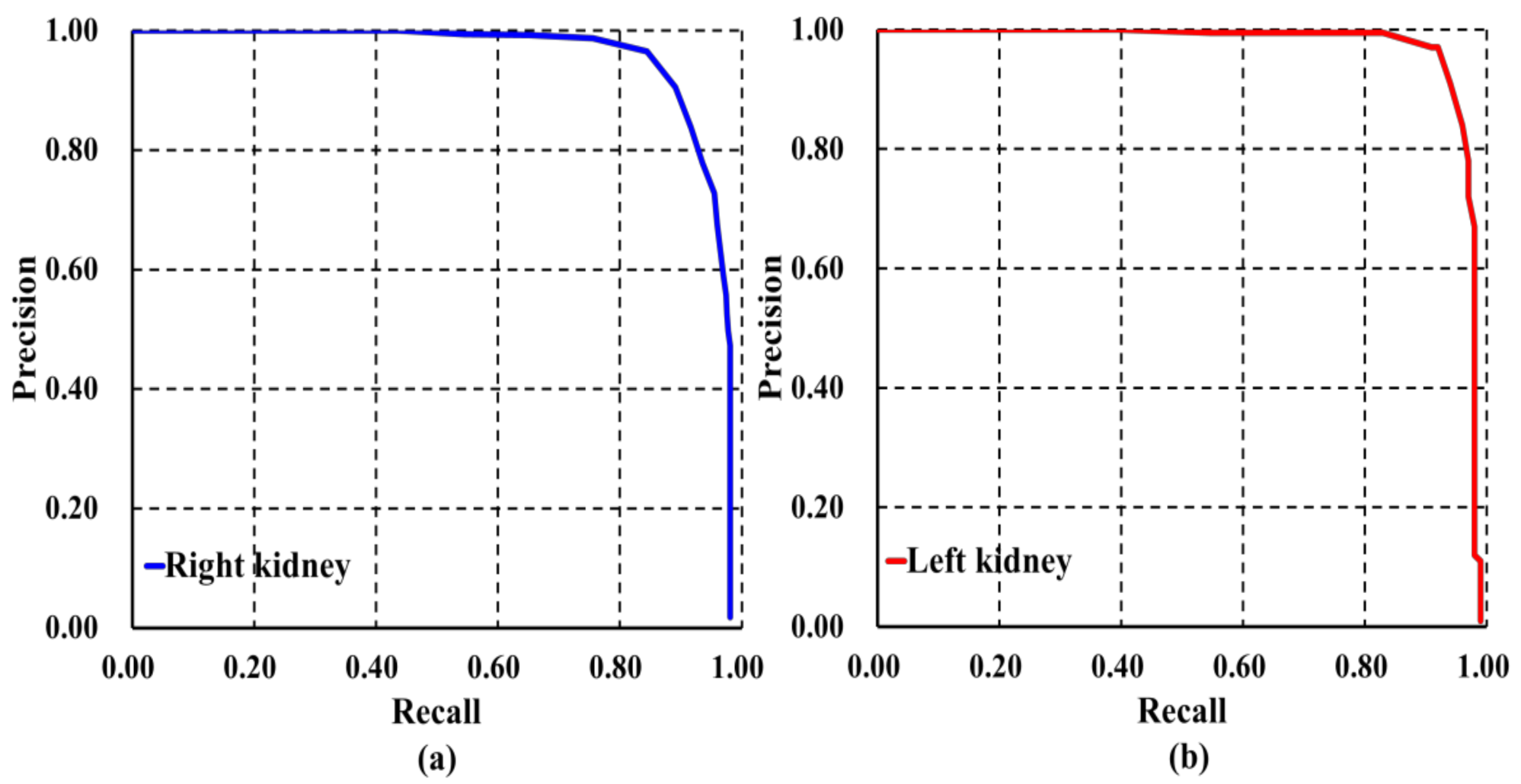
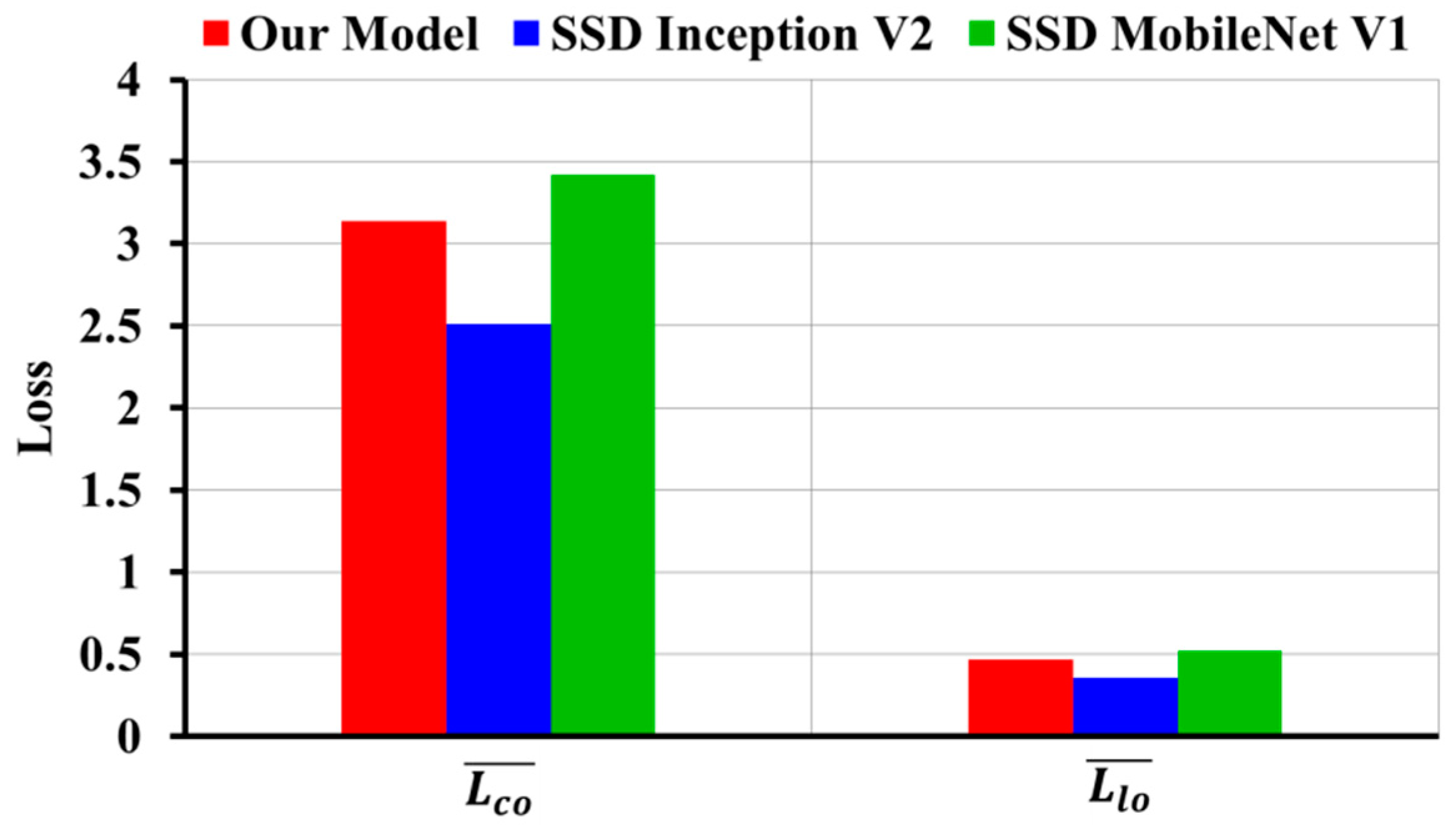
3.1. Evaluation Results of Image-Wise Testing
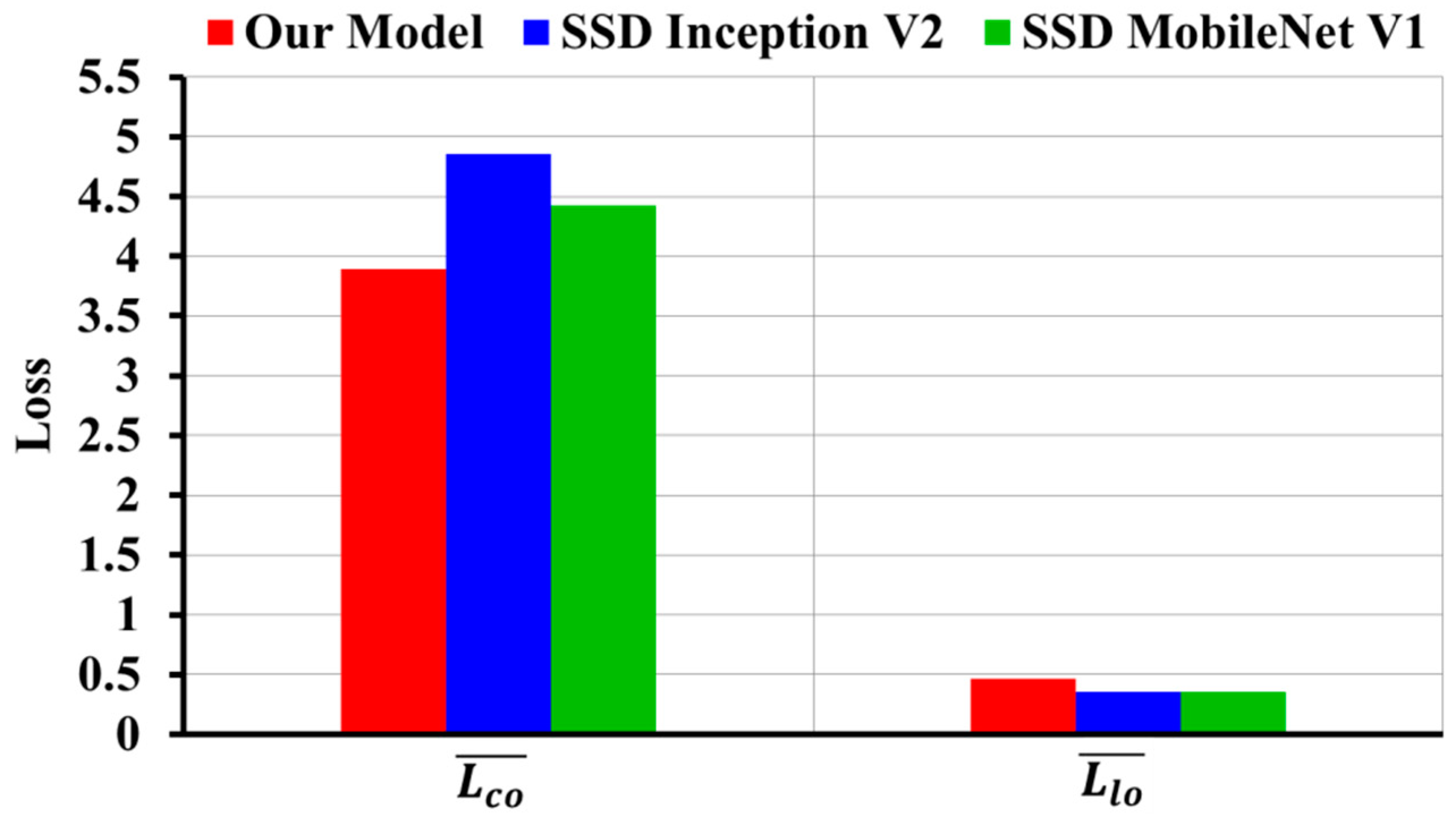
3.2. Evaluation Results of Subject-Wise Testing
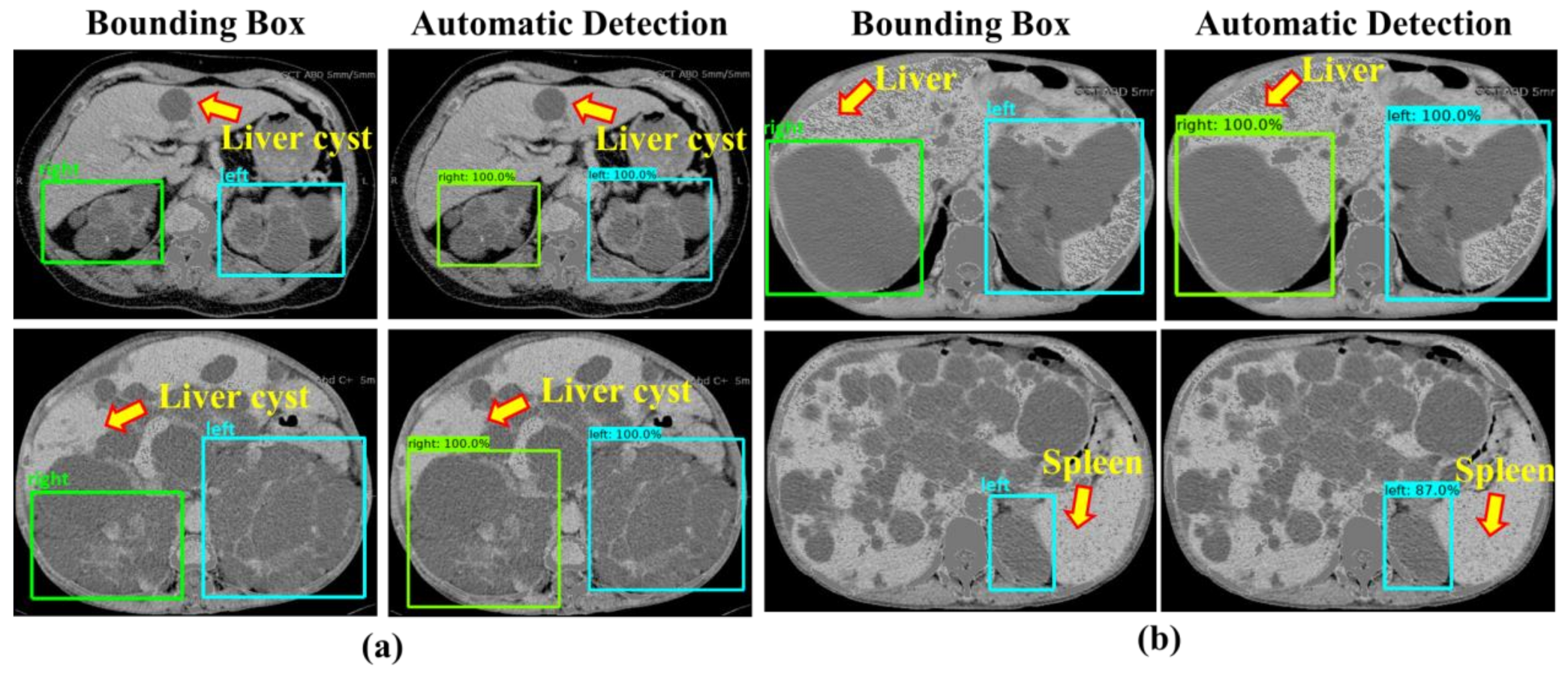
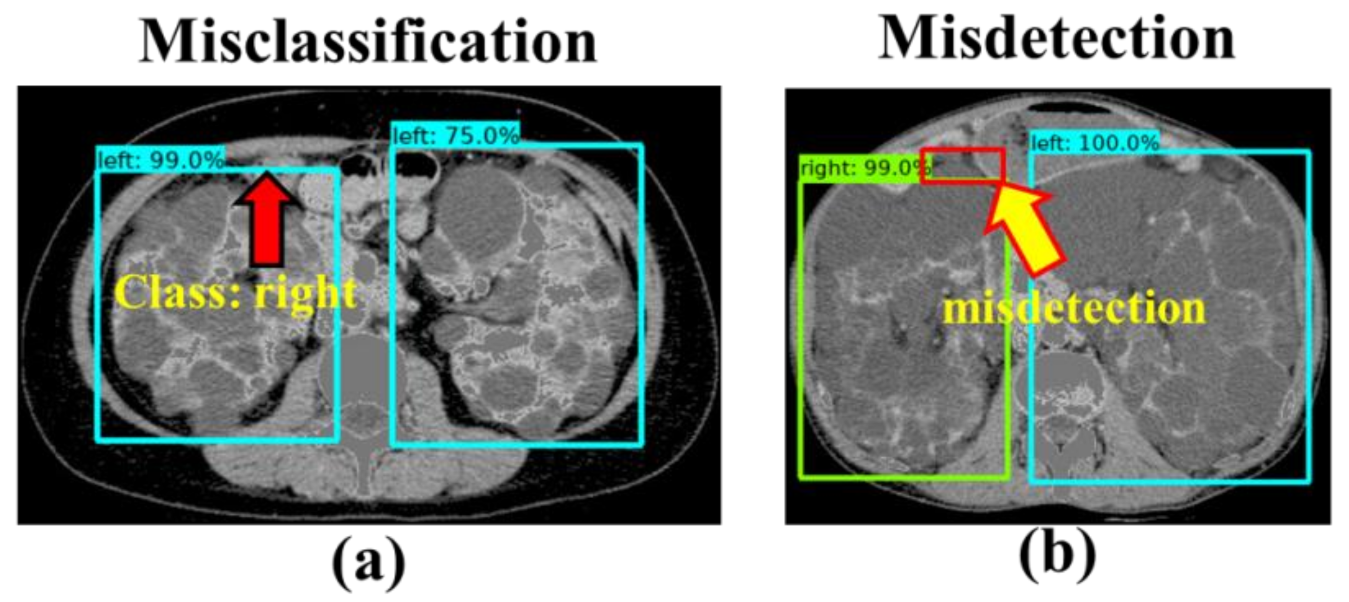
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lanktree, M.B.; Haghighi, A.; Guiard, E.; Iliuta, I.A.; Song, X.; Harris, P.C.; Paterson, A.D.; Pei, Y. Prevalence Estimates of Polycystic Kidney and Liver Disease by Population Sequencing. J. Am. Soc. Nephrol. 2018, 29, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Willey, C.J.; Blais, J.D.; Hall, A.K.; Krasa, H.B.; Makin, A.J.; Czerwiec, F.S. Prevalence of autosomal dominant polycystic kidney disease in the European Union. Nephrol. Dial. Transplant. 2017, 32, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Cornec-Le Gall, E.; Alam, A.; Perrone, R.D. Autosomal dominant polycystic kidney disease. Lancet 2019, 393, 919–935. [Google Scholar] [CrossRef]
- The US Food and Drug Administration. Total Kidney Volume Qualified as a Biomarker. Available online: https://www.raps.org/news-articles/news-articles/2016/9/total-kidney-volume-qualified-as-a-biomarker-by-fda-for-adpkd-trials?feed=Regulatory-Focus (accessed on 8 August 2020).
- Irazabal, M.V.; Rangel, L.J.; Bergstralh, E.J.; Osborn, S.L.; Harmon, A.J.; Sundsbak, J.L.; Bae, K.T.; Chapman, A.B.; Grantham, J.J.; Mrug, M.; et al. Imaging classification of autosomal dominant polycystic kidney disease: A simple model for selecting patients for clinical trials. J. Am. Soc. Nephrol. 2015, 26, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Chiang, S.F.; Sahoo, P.K.; Mohapatra, S.K.; You, J.F.; Onthoni, D.D.; Hung, H.Y.; Chiang, J.M.; Huang, Y.L.; Tsai, W.S. Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers 2019, 11, 2007. [Google Scholar] [CrossRef] [PubMed]
- Riaz, H.; Park, J.; Choi, H.; Kim, H.; Kim, J. Deep and Densely Connected Networks for Classification of Diabetic Retinopathy. Diagnostics 2020, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Pehrson, L.M.; Nielsen, M.B.; Lauridsen, C.A. Automatic Pulmonary Nodule Detection Applying Deep Learning or Machine Learning Algorithms to the LIDC-IDRI Database: A Systematic Review. Diagnostics 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of YOLO and grabcut algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Rupprecht, C.; Caroli, A.; Aparicio, M.C.; Remuzzi, A.; Baust, M.; Navab, N. Automatic Segmentation of Kidneys using Deep Learning for Total Kidney Volume Quantification in Autosomal Dominant Polycystic Kidney Disease. Sci. Rep. 2017, 7, 2049. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, D.; Georgescu, B.; Xu, D.; Comaniciu, D. Deep learning based automatic segmentation of pathological kidney in CT: Local versus global image context. In Deep Learning and Convolutional Neural Networks for Medical Image Computing; Springer: Berlin/Heidelberg, Germany, 2017; pp. 241–255. [Google Scholar]
- Kline, T.L.; Korfiatis, P.; Edwards, M.E.; Blais, J.D.; Czerwiec, F.S.; Harris, P.C.; King, B.F.; Torres, V.E.; Erickson, B.J. Performance of an Artificial Multi-observer Deep Neural Network for Fully Automated Segmentation of Polycystic Kidneys. J. Digit. Imaging 2017, 30, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Keshwani, D.; Kitamura, Y.; Li, Y. Computation of Total Kidney Volume from CT Images in Autosomal Dominant Polycystic Kidney Disease Using Multi-task 3D Convolutional Neural Networks. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 380–388. [Google Scholar]
- Brunetti, A.; Cascarano, G.D.; De Feudis, I.; Moschetta, M.; Gesualdo, L.; Bevilacqua, V. Detection and Segmentation of Kidneys from Magnetic Resonance Images in Patients with Autosomal Dominant Polycystic Kidney Disease. In Proceedings of the International Conference on Intelligent Computing, Nanchang, China, 3–6 August 2019; Springer: Cham, Switzerland, 2019; pp. 639–650. [Google Scholar]
- Bevilacqua, V.; Brunetti, A.; Cascarano, G.D.; Palmieri, F.; Guerriero, A.; Moschetta, M. A deep learning approach for the automatic detection and segmentation in autosomal dominant polycystic kidney disease based on magnetic resonance images. In Proceedings of the International Conference on Intelligent Computing, Wuhan, China, 15–18 August 2018; Springer: Cham, Switzerland, 2018; pp. 643–649. [Google Scholar]
- Bevilacqua, V.; Brunetti, A.; Cascarano, G.D.; Guerriero, A.; Pesce, F.; Moschetta, M.; Gesualdo, L. A comparison between two semantic deep learning frameworks for the autosomal dominant polycystic kidney disease segmentation based on magnetic resonance images. BMC Med. Inform. Decis. Mak. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Wang, X.; Lu, L.; Summers, R.M. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging (Bellingham) 2018, 5, 036501. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, X.G.; Liu, W.Y.; Latecki, L.J.; Wang, B.; Huang, J.Z. Weakly supervised mitosis detection in breast histopathology images using concentric loss. Med. Image Anal. 2019, 53, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, P.K.; Soltani, S.; Wong, A.K. A survey of thresholding techniques. Comput. Vis. Graph. Image Process. 1988, 41, 233–260. [Google Scholar] [CrossRef]
- Maragos, P. Tutorial on advances in morphological image processing and analysis. Opt. Eng. 1987, 26, 267623. [Google Scholar] [CrossRef]
- Suzuki, S. Topological structural analysis of digitized binary images by border following. Comput. Vis. Graph. Image Process. 1985, 30, 32–46. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Tzutalin, L. Git Code. Available online: https://github.com/tzutalin/labelImg (accessed on 8 January 2020).
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 21–37. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- He, Q.P.; Wang, J. Application of Systems Engineering Principles and Techniques in Biological Big Data Analytics: A Review. Processes 2020, 8, 951. [Google Scholar] [CrossRef]
- EL-Bana, S.; Al-Kabbany, A.; Sharkas, M. A Two-Stage Framework for Automated Malignant Pulmonary Nodule Detection in CT Scans. Diagnostics 2020, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.F.; Katiyar, S.; Keskar, A.G.; Bokde, N.D.; Geem, Z.W. Efficient Pneumonia Detection in Chest Xray Images Using Deep Transfer Learning. Diagnostics 2020, 10, 417. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rathod, V.; Sun, C.; Zhu, M.; Korattikara, A.; Fathi, A.; Fischer, I.; Wojna, Z.; Song, Y.; Guadarrama, S. Speed/accuracy trade-offs for modern convolutional object detectors. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7310–7311. [Google Scholar]
- Li, H.; Chaudhari, P.; Yang, H.; Lam, M.; Ravichandran, A.; Bhotika, R.; Soatto, S. Rethinking the Hyperparameters for Fine-tuning. arXiv 2020, arXiv:2002.11770. [Google Scholar]


| Characteristic | Mean ± SDs (Range) or Number | |
|---|---|---|
| Age at examination (yrs) | 54.59 ± 18.5 (27–88) | |
| Sex | Male | 52 |
| Female | 45 | |
| TKV (cm3) | 2734.33 ± 2312.45 (345.14–13,666.88) | |
| Detection Architectures | Class | Evaluation Metrics | |||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | ||
| Our Model | right | 0.90 (±0.06) | 0.92 (±0.07) | 0.82 (±0.02) | 0.86 (±0.04) |
| left | 0.91 (±0.06) | 0.92 (±0.06) | 0.84 (±0.06) | 0.88 (±0.05) | |
| Single Shot Detector (SSD) Inception V2 | right | 0.86 (±0.04) | 0.90 (±0.03) | 0.80 (±0.04) | 0.84 (±0.02) |
| left | 0.86 (±0.04) | 0.91 (±0.03) | 0.82 (±0.03) | 0.86 (±0.03) | |
| SSD MobileNet V1 | right | 0.73 (±0.1) | 0.75 (±0.01) | 0.72 (±0.1) | 0.72 (±0.09) |
| left | 0.71 (±0.1) | 0.81 (±0.06) | 0.66 (±0.1) | 0.71 (±0.1) | |
| Faster Region with Convolutional Neural Networks (R-CNN) NAS | right | 0.57 (±0.1) | 0.51 (±0.04) | 0.69 (±0.07) | 0.58 (±0.01) |
| left | 0.46 (±0.06) | 0.69 (±0.02) | 0.43 (±0.06) | 0.52 (±0.04) | |
| Faster R-CNN Inception ResNet V2 | right | 0.27 (±0.05) | 0.43 (±0.09) | 0.26 (±0.09) | 0.31 (±0.07) |
| left | 0.52 (±0.04) | 0.50 (±0.02) | 0.67 (±0.1) | 0.57 (±0.06) | |
| Region-FCN (R-FCN) ResNet 101 | right | 0.37 (±0.09) | 0.57 (±0.1) | 0.34 (±0.08) | 0.42 (±0.07) |
| left | 0.53 (±0.08) | 0.58 (±0.05) | 0.67 (±0.06) | 0.62 (±0.05) | |
| Detection Architectures | |||
|---|---|---|---|
| AP | mAP | ||
| Right | Left | Both Classes | |
| Our Model | 0.934 (±0.01) | 0.944 (±0.01) | 0.94 (±0.01) |
| SSD Inception V2 | 0.844 (±0.03) | 0.866 (±0.04) | 0.855 (±0.03) |
| SSD MobileNet V1 | 0.679 (±0.10) | 0.705 (±0.10) | 0.692 (±0.09) |
| Faster R-CNN NAS | 0.594 (±0.03) | 0.625 (±0.03) | 0.609 (±0.03) |
| Faster R-CNN Inception ResNet V2 | 0.491 (±0.08) | 0.568 (±0.06) | 0.53 (±0.07) |
| R-FCN ResNet 101 | 0.506 (±0.11) | 0.649 (±0.11) | 0.578 (±0.11) |
| Detection Architectures | Class | Evaluation Metrics | |||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall/Sensitivity | F1-Score | ||
| Our Model | right | 0.8 | 0.8 | 0.8 | 0.8 |
| left | 0.817 | 0.8 | 0.84 | 0.81 | |
| SSD Inception V2 | right | 0.434 | 0.552 | 0.512 | 0.531 |
| left | 0.511 | 0.582 | 0.632 | 0.606 | |
| SSD MobileNet V1 | right | 0.563 | 0.574 | 0.652 | 0.611 |
| left | 0.549 | 0.654 | 0.558 | 0.602 | |
| Faster R-CNN NAS | right | 0.483 | 0.394 | 0.728 | 0.511 |
| left | 0.248 | 0.549 | 0.232 | 0.326 | |
| Faster R-CNN Inception ResNet V2 | right | 0.268 | 0.291 | 0.266 | 0.278 |
| left | 0.513 | 0.464 | 0.694 | 0.556 | |
| R-FCN ResNet 101 | right | 0.3 | 0.464 | 0.245 | 0.332 |
| left | 0.529 | 0.572 | 0.676 | 0.620 | |
| Detection Architectures | Evaluation Metrics | ||
|---|---|---|---|
| AP | mAP | ||
| Right | Left | Both Classes | |
| Our Model | 0.80 | 0.852 | 0.824 |
| SSD Inception V2 | 0.42 | 0.497 | 0.458 |
| SSD MobileNet V1 | 0.567 | 0.536 | 0.551 |
| Faster R-CNN NAS | 0.48 | 0.549 | 0.515 |
| Faster R-CNN Inception ResNet V2 | 0.483 | 0.612 | 0.548 |
| R-FCN ResNet 101 | 0.474 | 0.681 | 0.577 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onthoni, D.D.; Sheng, T.-W.; Sahoo, P.K.; Wang, L.-J.; Gupta, P. Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images. Diagnostics 2020, 10, 1113. https://doi.org/10.3390/diagnostics10121113
Onthoni DD, Sheng T-W, Sahoo PK, Wang L-J, Gupta P. Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images. Diagnostics. 2020; 10(12):1113. https://doi.org/10.3390/diagnostics10121113
Chicago/Turabian StyleOnthoni, Djeane Debora, Ting-Wen Sheng, Prasan Kumar Sahoo, Li-Jen Wang, and Pushpanjali Gupta. 2020. "Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images" Diagnostics 10, no. 12: 1113. https://doi.org/10.3390/diagnostics10121113
APA StyleOnthoni, D. D., Sheng, T.-W., Sahoo, P. K., Wang, L.-J., & Gupta, P. (2020). Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images. Diagnostics, 10(12), 1113. https://doi.org/10.3390/diagnostics10121113







