Differential Diagnosis of Preeclampsia Based on Urine Peptidome Features Revealed by High Resolution Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Urine Sample Collection and Peptide Isolation
2.3. HPLC-MS/MS Analysis
2.4. Urinary Proteome Data Base Development
2.5. Data Analysis
3. Results
3.1. Urine Peptidome Analysis
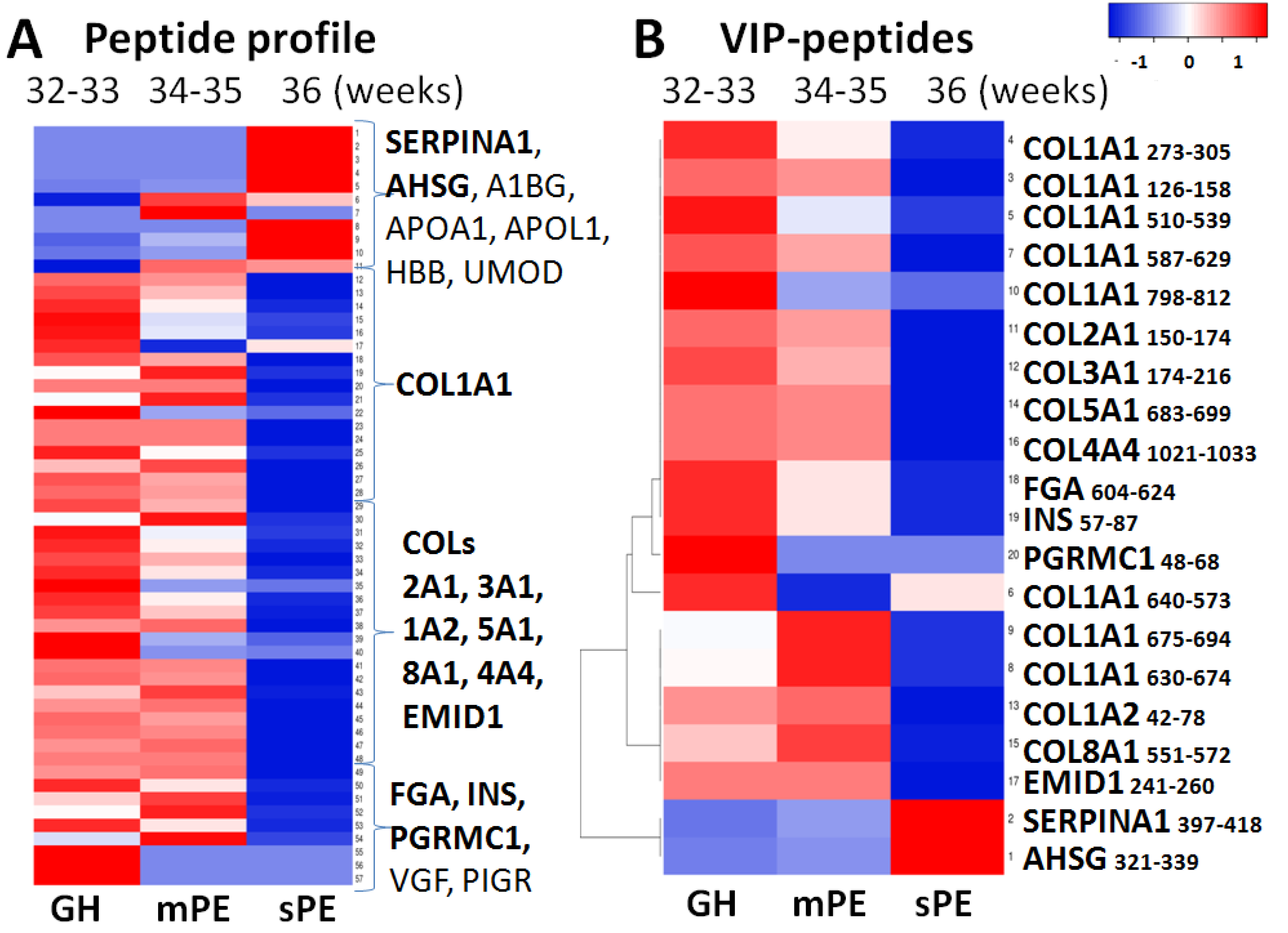
3.2. Peptide Groups
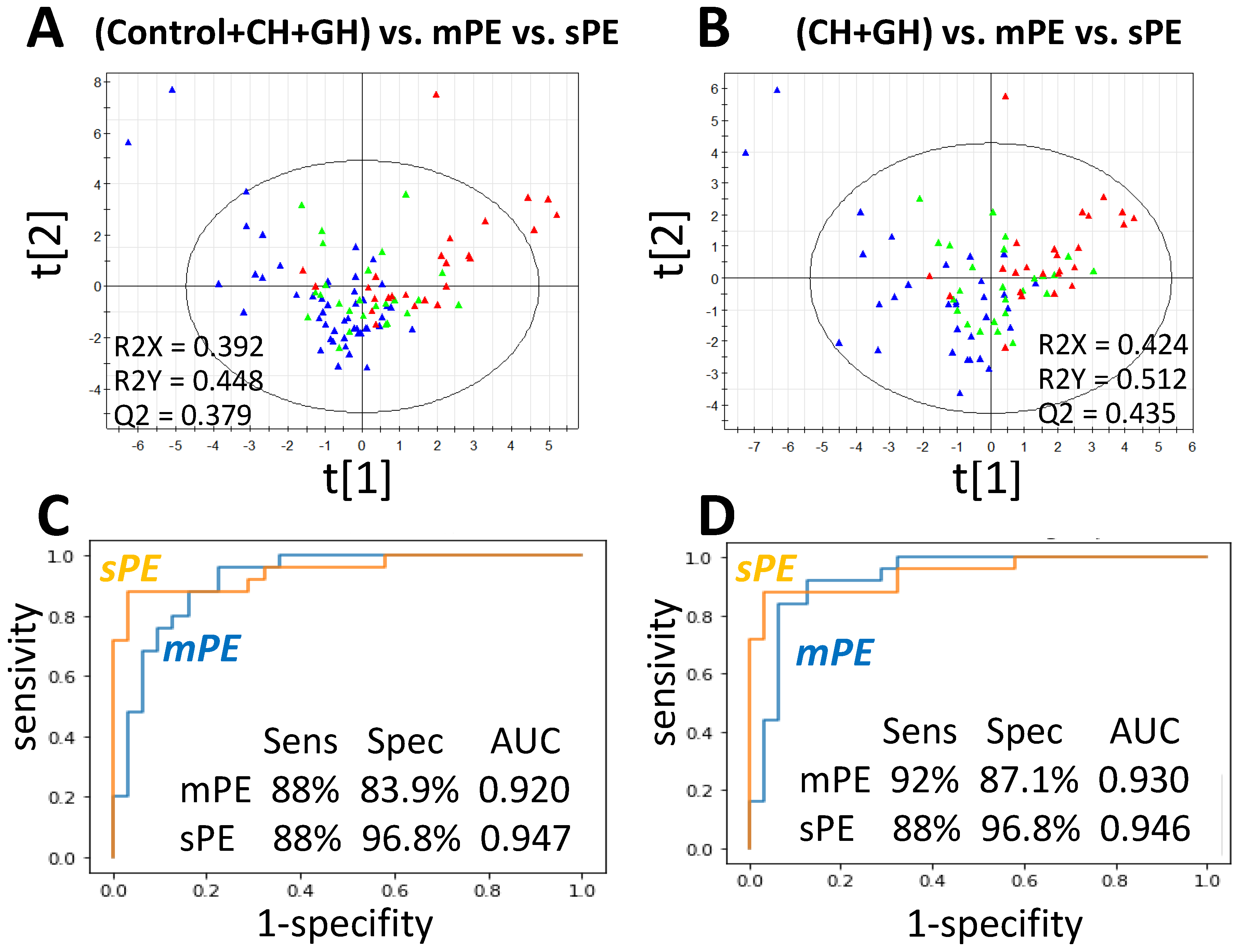
3.3. Predictive Performance and Model for PE Differentiation
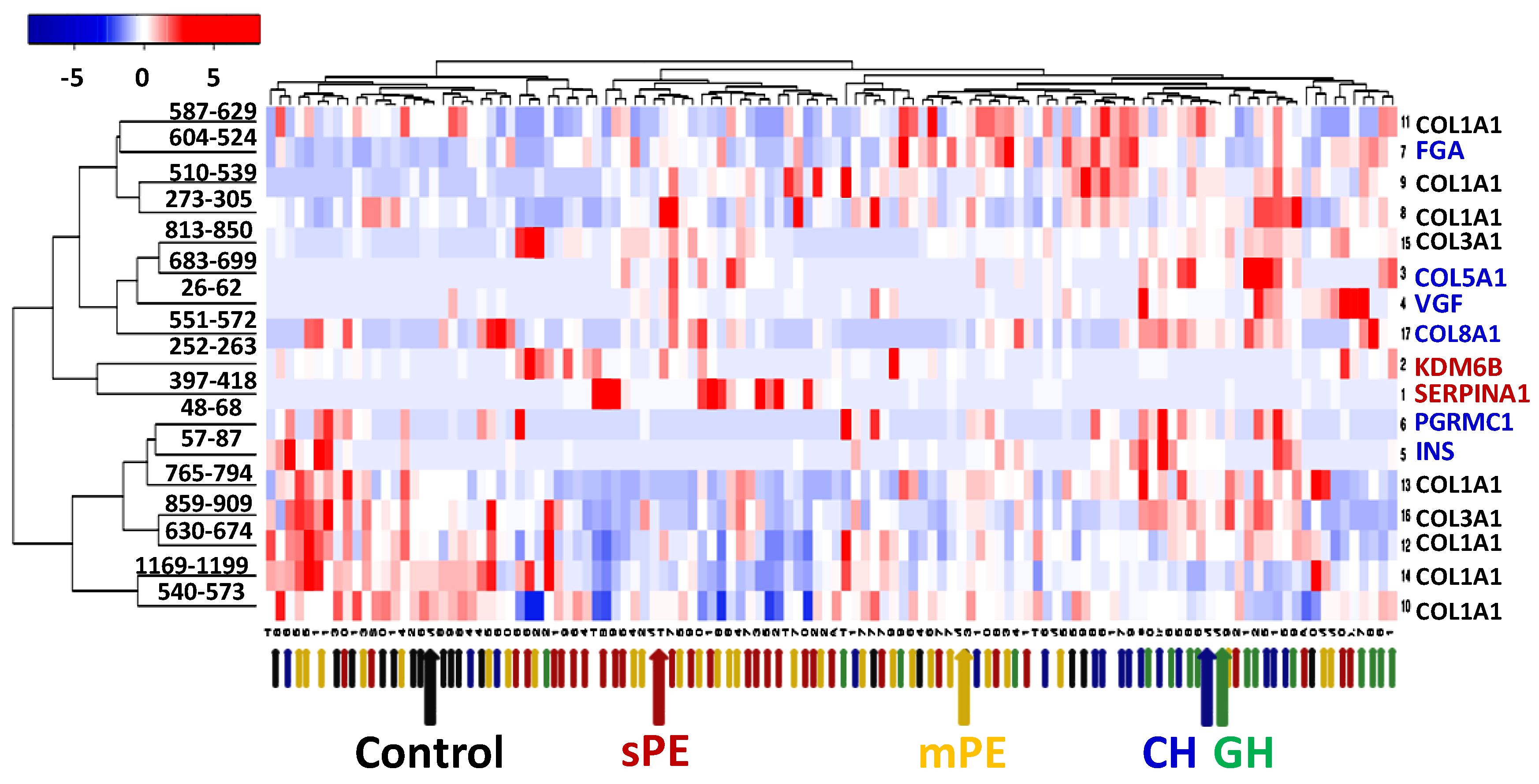
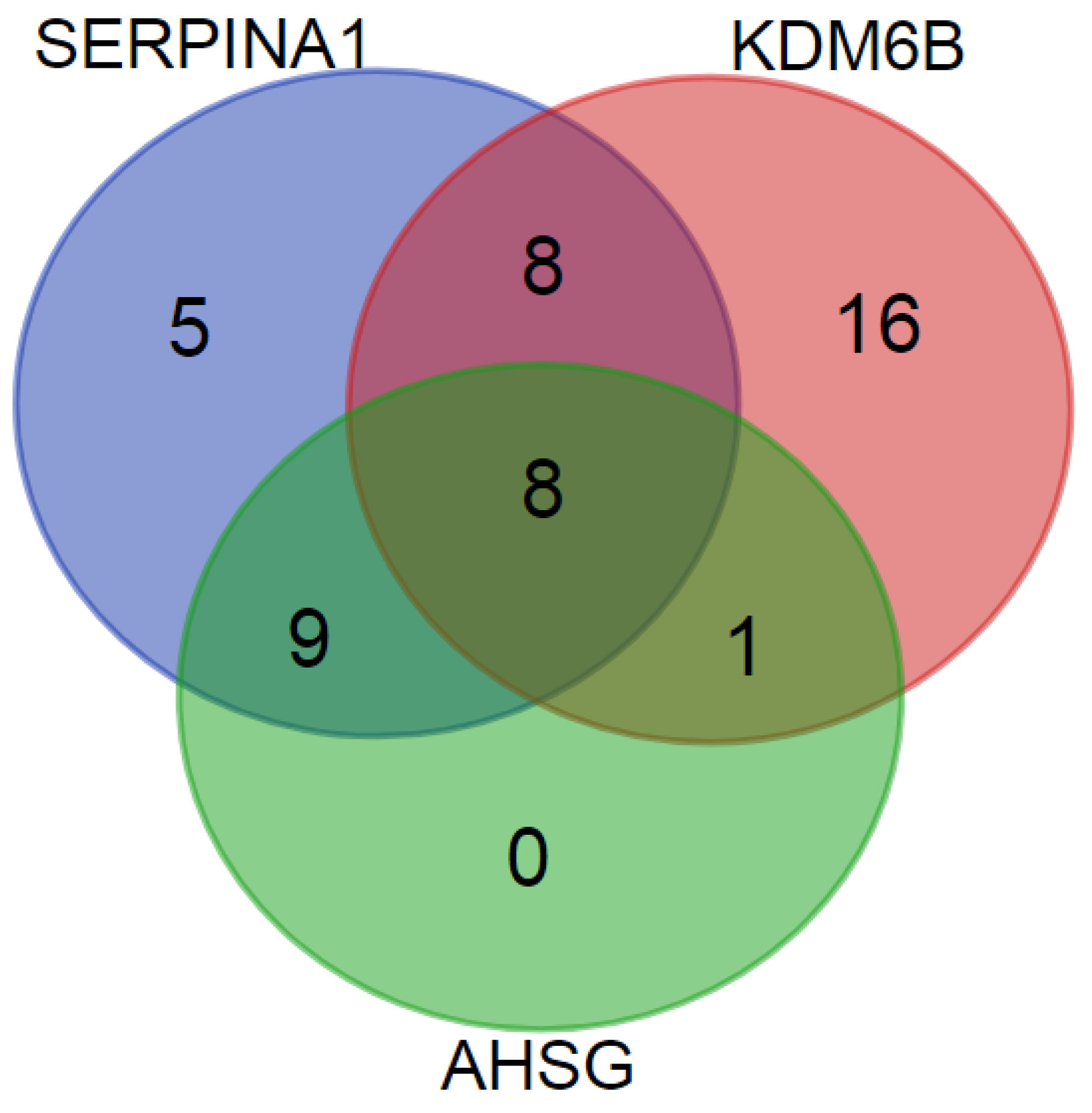
3.4. PE Markers
3.5. Samples from Patients with Complicated Diagnoses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Say, L.; Chou, D.; Gemmill, A.; Tuncalp, O.; Moller, A.B.; Daniels, J.; Gulmezoglu, A.M.; Temmerman, M.; Alkema, L. Global causes of maternal death: A WHO systematic analysis. Lancet. Glob. Health 2014, 2, e323–e333. [Google Scholar] [CrossRef]
- Fortunato, S.J.; Menon, R. Distinct molecular events suggest different pathways for preterm labor an premature rupture of membranes. Am. J. Obstet. Gynecol. 2001, 184, 1399–1405. [Google Scholar] [CrossRef]
- Bellamy, L.; Casas, J.P.; Hingorani, A.D.; Williams, D.J. Pre-eclampsia and risk of cardiovascular disease and cancer in later life: Systrmatic review and meta-analysis. BMJ 2007, 335, 974. [Google Scholar] [CrossRef]
- Backes, C.H.; Markham, K.; Moorehead, P.; Cordero, L.; Nankervis, C.A.; Giannone, P.J. Maternal preeclampsia and neonatal outcomes. J. Pregnancy 2011, 2011, 214365. [Google Scholar] [CrossRef]
- Brennan, L.J.; Morton, J.S.; Davidge, S.T. Vascular dysfunction in preeclampsia. Microcirculation 2014, 21, 4–14. [Google Scholar] [CrossRef]
- Buhimschi, I.A.; Nayeri, U.A.; Zhao, G.; Shook, L.L.; Pensalfini, A.; Funai, E.F.; Bernstein, I.M.; Glabe, C.G.; Buhimschi, C.S. Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci. Transl. Med. 2014, 6, 245ra92. [Google Scholar] [CrossRef]
- McCarthy, F.P.; Adetoba, A.; Gill, C.; Bramham, K.; Bertolaccini, M.; Burton, G.J.; Girardi, G.; Seed, P.T.; Poston, L.; Chappell, L.C. Urinary congophilia in women with hypertensive disorders of pregnancy and preexisting proteinuria or hypertension. Am. J. Obstet. Gynecol. 2016, 215, 464–e1. [Google Scholar] [CrossRef]
- Nagarajappa, C.; Rangappa, S.S.; Suryanarayana, R.; Balakrishna, S. Urinary congophilia in preeclampsia: Experience from a rural tertiary-care hospital in India. Pregnancy Hypertens. 2018, 13, 83–86. [Google Scholar] [CrossRef]
- Roberts, J.M.; Hubel, C.A. Oxidative stress in preeclampsia. Am. J. Obstet. Gynecol. 2004, 190, 1177–1178. [Google Scholar] [CrossRef]
- Borzychowski, A.M.; Sargent, I.L.; Redman, C.W. Inflammation and pre-eclampsia. Semin. Fetal Neonatal Med. 2006, 11, 309–316. [Google Scholar] [CrossRef]
- English, F.A.; Kenny, L.C.; McCarthy, F.P. Risk factors and effective management of preeclampsia. Integr. Blood Press. Control 2015, 8, 7–12. [Google Scholar] [CrossRef]
- Redman, C.W.; Sargent, I.L.; Staff, A.C. IFPA Senior Award Lecture: Making sense of pre-eclampsia—two placental causes of preeclampsia? Placenta 2014, 35, S20–S25. [Google Scholar] [CrossRef]
- Valdes, G.; Kaufmann, P.; Corthorn, J.; Erices, R.; Brosnihan, K.B.; Joyner-Grantham, J. Vasodilator factors in the systemic and local adaptations to pregnancy. Reprod. Biol Endocrinol. 2009, 7, 79. [Google Scholar] [CrossRef]
- Tanbe, A.F.; Khalil, R.A. Circulating and vascular bioactive factors during hypertension in pregnancy. Curr. Bioact. Compd. 2010, 6, 60–75. [Google Scholar] [CrossRef]
- Chen, J.; Khalil, R.A. Matrix matelloproteinases in normal pregnancy and preeclampsia. Prog. Mol. Biol. Transl. Sci. 2017, 148, 87–165. [Google Scholar] [CrossRef]
- Chelbi, S.T.; Vaiman, D. Genetic and epigenetic factors contribute to the onset of preeclampsia. Mol. Cell. Endocrinol. 2008, 282, 120–129. [Google Scholar] [CrossRef]
- Duckitt, K.; Harrington, D. Risk factors for pre-eclampsia at antenatal booking: Systematic review of controlled studies. BMJ 2005, 330, 565. [Google Scholar] [CrossRef]
- Rosenberg, T.J.; Garbers, S.; Lipkind, H.; Chiasson, M.A. Maternal obesity and diabetes as risk factors for adverse pregnancy outcomes: Differences among 4 racial/ethnic groups. Am. J. Public. Health 2005, 95, 1545–1551. [Google Scholar] [CrossRef]
- Nakajima, T.; Jorde, L.B.; Ishigami, T.; Umemura, S.; Emi, M.; Lalouel, J.M.; Inoue, I. Nucleotide diversity and haplotype structure of the human angiotensinogen gene in two populations. Am. J. Hum. Genet. 2002, 70, 108–123. [Google Scholar] [CrossRef]
- Ward, K.; Taylor, R.N. Genetic factors in the etiology of preeclampsia/eclampsia. In Chesley’s Hypertensive Disorders in Pregnancy, 4th ed.; Academic Press: Cambridge, MA, USA, 2015; Chapter 4; pp. 57–80. ISBN 978-0-12-407866-6. [Google Scholar] [CrossRef]
- Van Dijk, M.; Oudejans, C.B. STOX1: Key player in trophoblast dysfunction underlying early onset preeclampsia with growth retardation. J. Pregnancy 2011, 2011, 521826. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Dai, L.; Song, Y.; Wang, Y.; Zhou, B.; Zhou, R. Association between polymorphishs in CXCR2 gene and preeclampsia. Mol. Genet. Genomic. Med. 2019, 2019, e578. [Google Scholar] [CrossRef]
- Papazoglou, D.; Galazios, G.; Koukourakis, M.I.; Kontomanolis, E.N.; Maltezos, E. Association of −634G/C and 936C/T polymorphisms of the vascular endothelial growth factor with spontaneous preterm delivery. Acta Obstet. Gynecol. Scand. 2004, 83, 461–465. [Google Scholar] [PubMed]
- Alrahmani, L.; Willrich, M.A.V. The complement alternative pathway and preeclampsia. Curr. Hypertens. Rep. 2018, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Shimonovitz, S.; Hurwitz, A.; Dushnik, M.; Anteby, E.; Geva-Eldar, T.; Yagel, S. Developmental regulation of the expression of 72 and 92 kd type IV collagenases in human trophoblasts: A possible mechanism for control of trophoblast invasion. Am. J. Obstet. Gynecol. 1994, 171, 832–838. [Google Scholar] [CrossRef]
- Isaka, K.; Usuda, S.; Ito, H.; Sagawa, Y.; Nakamura, H.; Nishi, H.; Suzuki, Y.; Li, Y.F.; Takayama, M. Expression and activity of matrix metalloproteinase 2 and 9 in human trophoblasts. Placenta 2003, 24, 53–64. [Google Scholar] [CrossRef]
- Su, M.T.; Tsai, P.Y.; Tsai, H.L.; Chen, Y.C.; Kuo, P.L. miR-346 and miR-582-3p-regulated EG-VEGF expression and trophoblast invasion via matrix metalloproteinases 2 and 9. Biofactors 2017, 43, 210–219. [Google Scholar] [CrossRef]
- Montagnana, M.; Lippi, G.; Albiero, A.; Scevarolli, S.; Salvagno, G.L.; Franchi, M.; Guidi, G.C. Evaluation of metalloproteinases 2 and 9 and their inhibitors in physiologic and pre-eclamptic pregnancy. J. Clin. Lab. Anal. 2009, 23, 88–92. [Google Scholar] [CrossRef]
- Eleuterio, N.M.; Palei, A.C.; Rangel Machado, J.S.; Tanus-Santos, J.E.; Cavalli, R.C.; Sandrim, V.C. Positive correlations between circulating adiponectin and MMP2 in preeclampsia pregnant. Pregnancy Hypertens. 2015, 5, 205–208. [Google Scholar] [CrossRef]
- Chappell, L.C.; Duckworth, S.; Seed, P.T.; Griffin, M.; Myers, J.; Mackillop, L.; Simpson, N.; Waugh, J.; Anumba, D.; Kenny, L.C.; et al. Diagnostic accuracy of placental growth factor in women with suspected preeclampsia: A prospective multicenter study. Circulation 2013, 128, 2121–2131. [Google Scholar] [CrossRef]
- March, M.I.; Geahchan, C.; Wenger, J.; Raghuraman, N.; Berg, A.; Haddow, H.; McKeon, B.A.; Narcisse, R.; David, J.L.; Scott, J.; et al. Circulating Angiogenic Factors and the Risk of Adverse Outcomes among Haitian Women with Preeclampsia. PLoS ONE 2015, 10, e0126815. [Google Scholar] [CrossRef]
- Maynard, S.E.; Min, J.Y.; Merchan, J.; Lim, K.H.; Li, J.; Mondal, S.; Libermann, T.A.; Morgan, J.P.; Sellke, F.W.; Stillman, I.E.; et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Investig. 2003, 111, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Shixia, C.; Duan, T. First-Trimester Maternal Serum Levels of sFLT1, PGF and ADMA Predict Preeclampsia. PLoS ONE 2015, 10, e0124684. [Google Scholar] [CrossRef] [PubMed]
- Venkatesha, S.; Toporsian, M.; Lam, C.; Hanai, J.; Mammoto, T.; Kim, Y.M.; Bdolah, Y.; Lim, K.H.; Yuan, H.T.; Libermann, T.A.; et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat. Med. 2006, 12, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.J.; Lam, C.; Qian, C.; Yu, K.F.; Maynard, S.E.; Sachs, B.P.; Sibai, B.M.; Epstein, F.H.; Romero, R.; Thandhani, R.; et al. Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. N. Engl. J. Med. 2006, 355, 992–1005. [Google Scholar] [CrossRef]
- Ali, S.M.; Khalil, R.A. Genetic, immune and vasoactive factors in the vascular dysfunction associated with hypertension in pregnancy. Expert Opin. Ther. Targets 2015, 19, 1495–1515. [Google Scholar] [CrossRef]
- Shah, D.A.; Khalil, R.A. Bioactive factors in uteroplacental and systemic circulation link placental ischemia to generalized vascular dysfunction in hypertensive pregnancy and preeclampsia. Biochem. Pharmacol. 2015, 95, 211–226. [Google Scholar] [CrossRef]
- Chafetz, I.; Kuhnreich, I.; Sammar, M.; Tol, Y.; Gibor, Y.; Meiri, H.; Cuckle, H.; Wolf, M. First-trimester placental protein 13 screening for preeclampsia and intrauterine growth restriction. Am. J. Obstet. Gynecol. 2007, 197, e1–e35. [Google Scholar] [CrossRef]
- Spencer, C.A.; Allen, V.M.; Flowerdew, G.; Dooley, K.; Dodds, L. Low levels of maternal serum PAPP-A in early pregnancy and the risk of adverse outcomes. Prenat. Diagn. 2008, 28, 1029–1036. [Google Scholar] [CrossRef]
- Karumanchi, S.A. Biomarkers in preeclampsia. In Biomarkers of Kidney Disease, 2nd ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 555–594, Chapter 14. [Google Scholar] [CrossRef]
- Guo, H.-X.; Zhu, Y.-B.; Wu, C.-P.; Zhong, M.; Hu, S.-W. Potential urine biomarkers for gestational hypertension and preeclampsia. Mol. Med. Rep. 2019, 19, 2463–2470. [Google Scholar] [CrossRef]
- Buhimschi, I.A.; Zhao, G.; Funai, E.F.; Harris, N.; Sasson, I.E.; Bernstein, I.M.; Saade, G.R.; Buhimschi, C.S. Proteomic profiling of urine identifies specific fragments of SERPINA1 and albumin as biomarkers of preeclampsia. Am. J. Obstet. Gynecol. 2008, 199, 551–e1. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Y.; Jin, X.; Zhang, L.; Zhou, Y.; Niu, J.; Chen, J.; Gu, Y. Urinary proteomics analysis for renal injury in hypertensive disorders of pregnancy with iTRAQ labeling and LC-MS/MS. Proteom. Clin. Appl. 2011, 5, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Carty, D.M.; Siwy, J.; Brennand, J.E.; Zurbig, P.; Mullen, W.; Franke, J.; McCulloch, J.W.; Noth, R.A.; Chappell, L.C.; Mischak, H.; et al. Urinary proteomics for prediction of preeclampsia. Hypertension 2011, 57, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Kononikhin, A.S.; Starodubtseva, N.L.; Bugrova, A.E.; Shirokova, V.A.; Chagovets, V.V.; Indeykina, M.I.; Popov, I.A.; Kostyukevich, Y.I.; Vavina, O.V.; Muminova, K.T.; et al. An untargeted approach for the analysis of the urine peptidome of women with preeclampsia. J. Proteom. 2016, 149, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Park, J.S.; Norwitz, E.R.; Kim, S.M.; Kim, B.J.; Park, C.-W.; Jun, J.K.; Syn, H.C. Characterization of discriminatory urinary proteomic biomarkers for severe preeclampsia using SELDI-TOF mass spectrometry. J. Perinat. Med. 2011, 39, 391–396. [Google Scholar] [CrossRef]
- Kolialexi, A.; Mavreli, D.; Tounta, G.; Mavrou, A.; Papantoniou, N. Urine proteomic studies in preeclampsia. Proteom. Clin. Appl. 2015, 9, 501–506. [Google Scholar] [CrossRef]
- Levine, R.J.; Thadhani, R.; Qian, C.; Lam, C.; Lim, K.-H.; Yu, K.F.; Blink, A.L.; Sachs, B.P.; Epstein, F.H.; Sibai, B.M.; et al. Urinary placental growth factor and risk of preeclampsia. JAMA 2005, 293, 77–85. [Google Scholar] [CrossRef]
- Kononikhin, A.S.; Sergeeva, V.A.; Bugrova, A.E.; Indeykina, M.I.; Starodubtseva, N.L.; Chagovets, V.V.; Popov, I.A.; Frankevich, V.E.; Pedrioli, P.; Sukhikh, G.T.; et al. Methodology for Urine Peptidome Analysis Based on Nano-HPLC Coupled to Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Methods Mol. Biol. 2018, 1719, 311–318. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewsky, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Prot. Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Sergeeva, V.A.; Zakharova, N.V.; Bugrova, A.E.; Starodubtseva, N.L.; Indeykina, M.I.; Kononikhin, A.S.; Frankevich, V.E.; Nikolaev, E.N. The high-resolution mass spectrometry study of the protein composition of amyloid-like urine aggregates associated with preeclampsia. Eur. J. Mass. Spectrom. 2020, 26, 158–161. [Google Scholar] [CrossRef]
- Starodubtseva, N.; Nizyaeva, N.; Baev, O.; Bugrova, A.; Gapaeva, M.; Muminova, K.; Kononikhin, A.; Frankevich, V.; Nikolaev, E.; Sukhikh, G. SERPINA1 peptides in urine as a potential marker of preeclampsia severity. Int. J. Mol. Sci. 2020, 21, 914. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Totaro, M.G.; Prosperini, E.; Notarbartolo, S.; Testa, G.; Natoli, G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
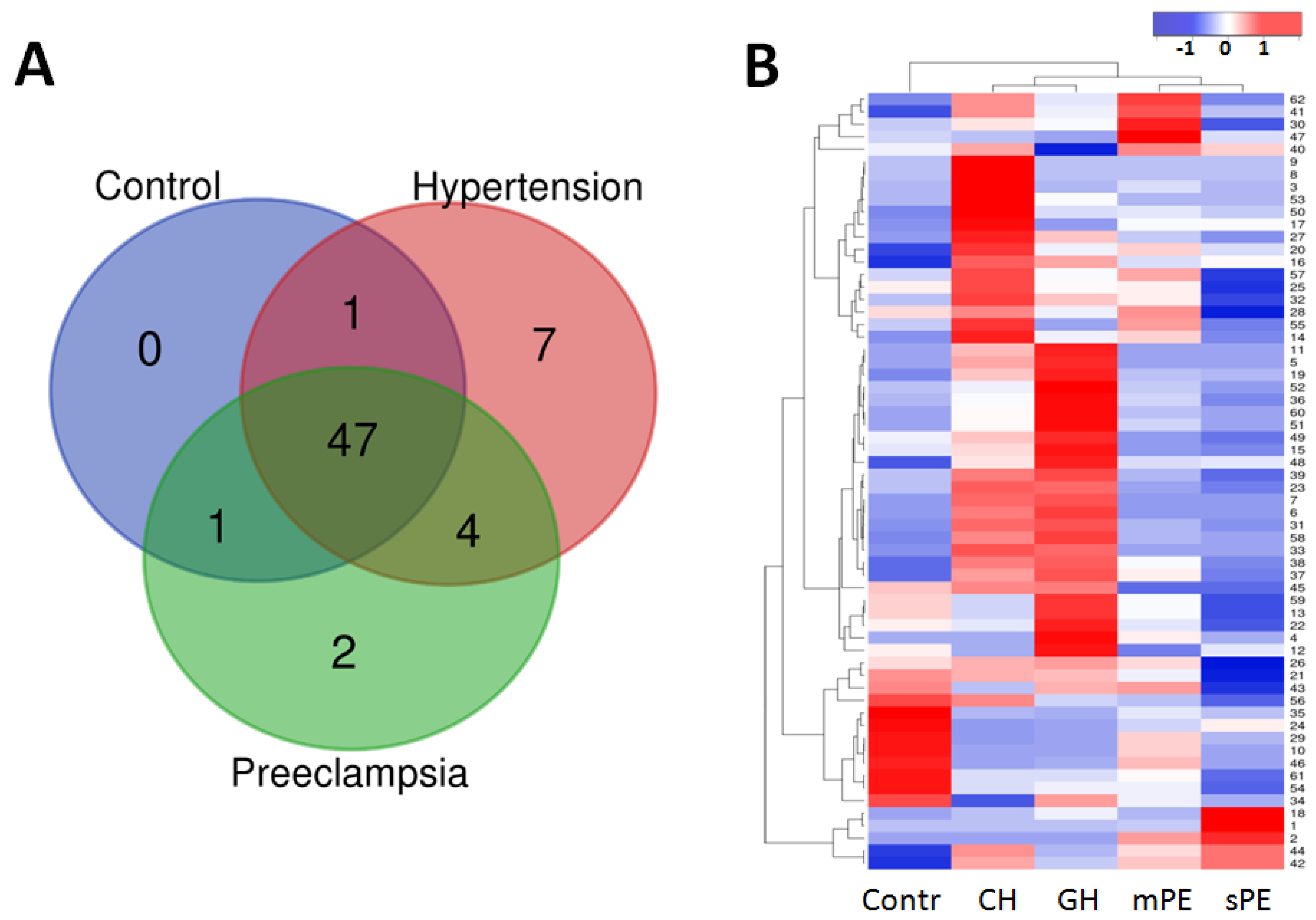

| Parameter | Control (n = 17) | CAH (n = 15) | GAH (n = 15) | Mild PE (n = 25) | Severe PE (n = 25) |
|---|---|---|---|---|---|
| Age (years) | 30.5 ± 4.1 | 31.8 ± 5 | 32.6 ± 4.2 | 28.5 ± 4.2 | 32.2 ± 4.5 |
| Height (cm) | 168.6 ± 4.8 | 167.4 ± 5 | 166.8 ± 7.1 | 164.6 ± 5.2 | 163.9 ± 5.8 |
| Weight (kg) | 70.1 ± 8.3 | 83 ± 15.5 | 76.5 ± 10.3 | 76.1 ± 14.9 | 74.2 ± 14.2 |
| BMI (kg/m2) | 24.7 ± 3.1 | 28.6 ± 3.4 | 27.2 ± 3.1 | 28.1 ± 5 | 27.6 ± 5.1 |
| Kidney disease | 2 (11.1%) | 2 (10.5%) | 4 (25%) | 2 (7.7%) | 4 (15.4%) |
| Previous PE | 1 (5.6%) | 6 (31.6%) | 1 (6.2%) | 0 (0%) | 6 (23.1%) |
| Primiparous | 5 (27.8%) | 7 (36.8%) | 5 (31.2%) | 16 (61.5%) | 11 (42.3%) |
| Start of hypertension (days) | - | 28 ± 9.1 | 29.2 ± 5.4 | 34.3 ± 5.1 | 28.2 ± 8.2 |
| Start of proteinuria (days) | - | 36.5 ± 0.7 | 33.5 ± 0.2 | 36.7 ± 2.1 | 32 ± 4.6 |
| sFlt-1/PLGF | 6.7 ± 1.8 | 33.4 ± 31.1 | 67.8 ± 78.1 | 229.8 ± 209.6 | 346.3 ± 281.9 |
| Delivery (weeks) | 39 ± 1.1 | 37.8 ± 1.9 | 37.6 ± 2.2 | 37.9 ± 1.6 | 33.6 ± 4.4 |
| Maximal SP | 117.2 ± 5.5 | 143.1 ± 14.4 | 146.7 ± 10.9 | 151.5 ± 12.6 | 157.7 ± 11.3 |
| Maximal DP | 77.8 ± 3.9 | 90.9 ± 7.3 | 94.7 ± 8.9 | 96.0 ± 6.9 | 100.8 ± 7.5 |
| Maximal Pu (g/l) | 0 ± 0 | 0.1 ± 0.1 | 0.1 ± 0.1 | 1.2 ± 1.0 | 2.3 ± 1.6 |
| LDH | 393.3 ± 0 | 327.6 ± 14.4 | 366.2 ± 33.8 | 334.5 ± 91.0 | 445.8 ± 117.1 |
| ALT | 13.8 ± 2.2 | 15 ± 6.6 | 17.9 ± 7 | 24.6 ± 28.7 | 36.1 ± 20.1 |
| AST | 17.3 ± 3.4 | 18.9 ± 4.6 | 16.8 ± 4.8 | 25.0 ± 12.2 | 33.9 ± 15.7 |
| ALP | 129.7 ± 3.4 | 201 ± 156.3 | 164.3 ± 68.3 | 168.5 ± 47 | 183.2 ± 89.2 |
| Platelet count | 217.3 ± 56.5 | 244.1 ± 73.4 | 251.5 ± 55.2 | 218.8 ± 67.8 | 199.5 ± 77.2 |
| UAPI | 0.9 ± 0 | 0.9 ± 0.3 | 1 ± 0.3 | 1.4 ± 0.6 | 2.0 ± 1.0 |
| UMAPI | 0.9 ± 0 | 1.1 ± 0.4 | 1 ± 0.3 | 1.3 ± 0.5 | 1.6 ± 1.3 |
| MCAPI | 1.6 ± 0 | 2.8 ± 0.7 | 1.8 ± 0.3 | 2.6 ± 1.2 | 2.8 ± 1.1 |
| IUGR | 0 (0%) | 0 (0%) | 2 (12.5%) | 6 (23.1%) | 17 (65.4%) |
| Child weight (g) | 3361.8 ± 505.7 | 3035.3 ± 518.6 | 2961 ± 562.5 | 2768.3 ± 540.5 | 1783.3 ± 834.4 |
| Child height (cm) | 51.4 ± 2.4 | 50.5 ± 2.6 | 49.9 ± 3.6 | 48.8 ± 2.6 | 40.9 ± 8 |
| Apgar 1 min | 8.1 ± 0.3 | 7.9 ± 0.3 | 7.8 ± 0.5 | 7.7 ± 0.5 | 6.6 ± 1.7 |
| Apgar 5 min | 9.1 ± 0.4 | 8.7 ± 0.6 | 8.6 ± 0.6 | 8.7 ± 0.5 | 7.7 ± 1.3 |
| Originating Protein | Number of Peptides | Number of Samples a | Diagnostic Groups b,c |
|---|---|---|---|
| COL1A1 | 196 | 127 | All groups |
| COL3A1 | 80 | 127 | All groups |
| COL1A2 | 10 | 110 | All groups |
| UMOD | 10 | 102 | All groups |
| FGA | 9 | 121 | All groups |
| COL18A1 | 8 | 122 | Contr, CH, GH, mPE, sPE |
| KISS1 | 4 | 58 | CH, GH, mPE |
| COL4A4 | 3 | 74 | Contr, CH, GH, mPE |
| FGB | 2 | 90 | Contr, CH, GH, mPE |
| COL2A1 | 2 | 74 | Contr, CH, GH, mPE |
| EMID1 | 2 | 74 | Contr, CH, GH, mPE |
| COL5A1 | 2 | 48 | CH, GH, mPE |
| COL8A1 | 2 | 43 | CH, GH |
| COL15A1 | 1 | 74 | Contr, CH, GH, mPE |
| COL17A1 | 1 | 74 | Contr, CH, GH, mPE |
| FXYD2 | 1 | 59 | Contr, mPE |
| PGRMC1 | 1 | 49 | CH |
| VGF | 1 | 49 | CH,GH |
| INS | 1 | 47 | CH,GH |
| KDM6B | 1 | 42 | sPE |
| PIGR | 1 | 31 | GH |
| Originating Protein | Number of Samples * | Intersection in Venn Diagram Number of Peptide Groups | |||||
|---|---|---|---|---|---|---|---|
| Core | Control/ CH+GH | Control/ PE | CH+GH/ PE | CH+GH | PE | ||
| Intersection | 47 | 1 | 1 | 4 | 7 | 2 | |
| UMOD | 118 (102) | 1 | - | - | - | - | - |
| KISS1 | 85 (58) | 1 | - | - | - | - | - |
| EMID1 | 103 (74) | 1 | - | - | - | - | - |
| FGA | 122 (121) | 1 | - | - | - | - | - |
| FGB | 95 (90) | 1 | - | - | - | - | - |
| COL2A1 | 106 (74) | 1 | - | - | - | - | - |
| COL8A1 | 93 (43) | 1 | - | - | - | - | - |
| COL15A1 | 92 (74) | 1 | - | - | - | - | - |
| COL17A1 | 93 (74) | 1 | - | - | - | - | - |
| COL3A1 | 127 (127) | 14 | 1 | - | - | - | - |
| COL1A1 | 127 (127) | 16 | - | - | 1 | - | - |
| COL18A1 | 125 (122) | 2 | - | - | 1 | - | - |
| COL1A2 | 114 (110) | 5 | - | - | - | 1 | - |
| COL4A4 | 106 (74) | 1 | - | - | - | 1 | - |
| FXYD2 | 59 (59) | - | - | 1 | - | - | - |
| PIGR | 57 (31) | - | - | - | 1 | - | - |
| COL5A1 | 53 (48) | - | - | - | 1 | 1 | - |
| COL5A2 | 48 (-) | - | - | - | - | 1 | - |
| INS | 50 (47) | - | - | - | - | 1 | - |
| VGF | 52 (49) | - | - | - | - | 1 | - |
| PGRMC1 | 51 (49) | - | - | - | - | 1 | - |
| KDM6B | 62 (42) | - | - | - | - | - | 1 |
| SERPINA1 | 47 (-) | - | - | - | - | - | 1 |
| Peptide Group | Start-End Position | Originating Protein | Number of Samples | Other Studies |
|---|---|---|---|---|
| GAAGEPGKAGERGVPGPPGAVGPAGKDGEAGAQGPPGPAGPAG | 587–629 | COL1A1 | 114 | |
| EAEDLQVGQVELGGGPGAGSLQPLALEGSLQ | 57–87 | INS | 50 | |
| LMIEQNTKSPLFMGKVVNPTQK | 397–418 | SERPINA1 | 47 | [42,45] |
| ERGSPGPAGPKGSPGEAGRPGEAGLPGAKG | 510–539 | COL1A1 | 98 | [44] |
| LLGPKGPPGPPGPPGVT | 683–699 | COL5A1 | 39 | |
| GRDGEPGTPGNPGPPGPPGPPGPPG | 150–174 | COL2A1 | 106 | |
| GPAGPPGPPGPPGTSGHPGSPGSPGYQGPPGEPGQAGPSGPPG | 174–216 | COL3A1 | 113 | |
| GQPGPPGPPGPPG | 1021–1033 | COL4A4 | 49 | |
| AGPPGRDGIPGQPGLPGPPGPPGPPGPPGLGGN | 126–158 | COL1A1 | 116 | |
| GPQGQPGLPGPPGPPGPPGPPA | 551–572 | COL8A1 | 93 | |
| MGVVSLGSPSGEVSHPRKT | 321–339 | AHSG | 26 | |
| GDQPAASGDSDDDEPPPLPRL | 48–68 | PGRMC1 | 51 | |
| ADEAGSEADHEGTHSTKRGHAKSRPV | 604–624 | FGA | 122 | [44] |
| ERGEQGPAGSPGFQGLPGPAGPPGEAGKPGEQGVPGDLGAPGPSG | 630–674 | COL1A1 | 127 | |
| VKGERGSPGGPGAAGFPGARGLPGPPGSNGNPGPPGPSGSPGKDGPPGPAG | 859–909 | COL3A1 | 125 | |
| GERGPPGPPGRDGEDGPTGPPGPPGPPGPPGLGGNFA | 42–78 | COL1A2 | 100 | |
| LTGPIGPPGPAGAPGDKGESGPSGPAGPTG | 765–794 | COL1A1 | 110 | [44] |
| PPPPPPPPPPPP | 252–263 | KDM6B | 62 | |
| LDGAKGDAGPAGPKGEPGSPGENGAPGQMGPRG | 273–305 | COL1A1 | 114 | |
| PGERGPPGPPGPPGPPGPPAP | 241–260 | EMID1 | 103 | |
| LTGSPGSPGPDGKTGPPGPAGQDGRPGPPGPPGA | 540–573 | COL1A1 | 127 | |
| APGDRGEPGPPGPAG | 798–812 | COL1A1 | 126 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kononikhin, A.S.; Zakharova, N.V.; Sergeeva, V.A.; Indeykina, M.I.; Starodubtseva, N.L.; Bugrova, A.E.; Muminova, K.T.; Khodzhaeva, Z.S.; Popov, I.A.; Shao, W.; et al. Differential Diagnosis of Preeclampsia Based on Urine Peptidome Features Revealed by High Resolution Mass Spectrometry. Diagnostics 2020, 10, 1039. https://doi.org/10.3390/diagnostics10121039
Kononikhin AS, Zakharova NV, Sergeeva VA, Indeykina MI, Starodubtseva NL, Bugrova AE, Muminova KT, Khodzhaeva ZS, Popov IA, Shao W, et al. Differential Diagnosis of Preeclampsia Based on Urine Peptidome Features Revealed by High Resolution Mass Spectrometry. Diagnostics. 2020; 10(12):1039. https://doi.org/10.3390/diagnostics10121039
Chicago/Turabian StyleKononikhin, Alexey S., Natalia V. Zakharova, Viktoria A. Sergeeva, Maria I. Indeykina, Natalia L. Starodubtseva, Anna E. Bugrova, Kamila T. Muminova, Zulfia S. Khodzhaeva, Igor A. Popov, Wenguang Shao, and et al. 2020. "Differential Diagnosis of Preeclampsia Based on Urine Peptidome Features Revealed by High Resolution Mass Spectrometry" Diagnostics 10, no. 12: 1039. https://doi.org/10.3390/diagnostics10121039
APA StyleKononikhin, A. S., Zakharova, N. V., Sergeeva, V. A., Indeykina, M. I., Starodubtseva, N. L., Bugrova, A. E., Muminova, K. T., Khodzhaeva, Z. S., Popov, I. A., Shao, W., Pedrioli, P., Shmakov, R. G., Frankevich, V. E., Sukhikh, G. T., & Nikolaev, E. N. (2020). Differential Diagnosis of Preeclampsia Based on Urine Peptidome Features Revealed by High Resolution Mass Spectrometry. Diagnostics, 10(12), 1039. https://doi.org/10.3390/diagnostics10121039








