Phenotypic and Genotypic Analysis of Resistant Helicobacter pylori Strains Isolated from Children with Gastrointestinal Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Data
2.2. Detection of H. pylori Genomic DNA Fragments
2.3. Classical Sequencing of H. pylori DNA Fragments
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malfertheiner, P.; Venerito, M.; Schulz, C. Helicobacter pylori Infection: New Facts in Clinical Management. Curr. Treat. Opt. Gastroenterol. 2018, 16, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Lin, Y.; Kikuchi, S. Helicobacter pylori Infection in Children and Adolescents. Adv. Exp. Med. Biol. 2019, 1149, 107–120. [Google Scholar] [PubMed]
- Kori, M.; Daugule, I.; Urbonas, V. Helicobacter pylori and Some Aspects of Gut Microbiota in Children. Helicobacter 2018, 23, e12524. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Peek, R.M. Helicobacter pylori, Cancer, and the Gastric Microbiota. Adv. Exp. Med. Biol. 2016, 908, 393–408. [Google Scholar] [PubMed]
- Roka, K.; Roubani, A.; Stefanaki, K.; Panayotou, I.; Roma, E.; Chouliaras, G. The Prevalence of Helicobacter pylori Gastritis in Newly Diagnosed Children with Inflammatory Bowel Disease. Helicobacter 2014, 19, 400–405. [Google Scholar]
- Lupu, V.V.; Ignat, A.; Ciubotariu, G.; Ciubară, A.; Moscalu, M.; Burlea, M. Helicobacter pylori Infection and Gastroesophageal Reflux in Children. Dis. Esophagus 2016, 29, 1007–1012. [Google Scholar] [PubMed]
- Yucel, O. Interactions between Helicobacter pylori and Gastroesophageal Reflux Disease. Esophagus 2019, 16, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.L.; Koletzko, S.; Goodman, K.; Bontems, P.; Cadranel, S.; Casswall, T.; Czinn, S.; Gold, B.D.; Guarner, J.; Elitsur, Y.; et al. Joint ESPGHAN/NASPGHAN Guidelines for the Management of Helicobacter pylori in Children and Adolescents (Update 2016). J. Pediatr. Gastroenterol. Nutr. 2017, 64, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, V.; Zullo, A.; Hassan, C.; Giorgio, F.; Rosania, R.; Ierardi, E. Mechanisms of Helicobacter pylori Antibiotic Resistance: An Updated Appraisal. World J. Gastrointest. Pathophysiol. 2011, 2, 35–41. [Google Scholar]
- De Francesco, V.; Margiotta, M.; Zullo, A.; Hassan, C.; Troiani, L.; Burattini, O.; Stella, F.; Di Leo, A.; Russo, F.; Marangi, S.; et al. Clarithromycin-Resistant Genotypes and Eradication of Helicobacter pylori. Ann. Intern. Med. 2006, 144, 94–100. [Google Scholar] [CrossRef]
- Martínez-Júlvez, M.; Rojas, A.L.; Olekhnovich, I.; Angarica, V.E.; Hoffman, P.S.; Sancho, J. Structure of RdxA—An Oxygen-Insensitive Nitroreductase Essential for Metronidazole Activation in Helicobacter pylori. FEBS J. 2012, 279, 4306–4317. [Google Scholar] [PubMed]
- Hoffman, P.S.; Goodwin, A.; Johnsen, J.; Magee, K.; Van Zanten, S.J.O.V. Metabolic Activities of Metronidazole-Sensitive and -Resistant Strains of Helicobacter pylori: Repression of Pyruvate Oxidoreductase and Expression of Isocitrate Lyase Activity Correlate with Resistance. J. Bacteriol. 1996, 178, 4822–4829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gościniak, G.; Biernat, M.; Grabińska, J.; Bińkowska, A.; Poniewierka, E.; Iwańczak, B. The Antimicrobial Susceptibility of Helicobacter pylori Strains Isolated from Children and Adults with Primary Infection in the Lower Silesia Region, Poland. Polish J. Microbiol. 2014, 63, 57–61. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Mégraud, F.; Lopez-Brea, M.; Andersen, L. European Multicentre Survey of In Vitro Antimicrobial Resistance in Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 820–823. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Broutet, N.; Cantagrel, A.; Andersen, L.; Alarcon, T.; López-Brea, M.; Mégraud, F. Comparison of the E test and Agar Dilution Method for Antimicrobial Suceptibility Testing of Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 549–552. [Google Scholar]
- Bińkowska, A.; Biernat, M.M.; Łaczmański, Ł.; Gościniak, G. Molecular Patterns of Resistance Among Helicobacter pylori Strains in South-Western Poland. Front. Microbiol. 2018, 9, 3154. [Google Scholar] [CrossRef]
- Hanafi, A.; Lee, W.C.; Loke, M.F.; Teh, X.; Shaari, A.; Dinarvand, M.; Lehours, P.; Mégraud, F.; Ruey Leow, A.H.; Vadivelu, J.; et al. Molecular and Proteomic Analysis of Levofloxacin and Metronidazole Resistant Helicobacter pylori. Front. Microbiol. 2016, 7, 2015. [Google Scholar] [CrossRef]
- Zabala Torrres, B.; Lucero, Y.; Lagomarcino, A.J.; Orellana-Manzano, A.; George, S.; Torres, J.P.; O’Ryan, M. Review: Prevalence and Dynamics of Helicobacter pylori Infection during Childhood. Helicobacter 2017, 22, e12399. [Google Scholar]
- Iwańczak, B.M.; Buchner, A.M.; Iwańczak, F. Clinical Differences of Helicobacter pylori Infection in Children. Adv. Clin. Exp. Med. 2017, 26, 1131–1136. [Google Scholar] [CrossRef]
- Koletzko, S.; Richy, F.; Bontems, P.; Crone, J.; Kalach, N.; Monteiro, M.L.; Gottrand, F.; Celinska-Cedro, D.; Roma-Giannikou, E.; Orderda, G.; et al. Prospective Multicentre Study on Antibiotic Resistance of Helicobacter pylori Strains Obtained from Children Living in Europe. Gut 2006, 55, 1711–1716. [Google Scholar] [CrossRef]
- Shu, X.; Yin, G.; Liu, M.; Peng, K.; Zhao, H.; Jiang, M. Antibiotics Resistance of Helicobacter pylori in Children with Upper Gastrointestinal Symptoms in Hangzhou, China. Helicobacter 2018, 23, e12481. [Google Scholar] [CrossRef] [PubMed]
- Dargiene, G.; Kupcinskas, J.; Jonaitis, L.; Vezbavicius, M.; Kadusevicius, E.; Kupcinskiene, E.; Frandsen, T.H.; Kucinskiene, R.; Kupcinskas, L.; Andersen, L.P. Primary Antibiotic Resistance of Helicobacter pylori Strains among Adults and Children in a Tertiary Referral Centre in Lithuania. APMIS 2018, 126, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Boyanova, L.; Gergova, G.; Evstatiev, I.; Spassova, Z.; Kandilarov, N.; Yaneva, P.; Markovska, R.; Mitov, I. Helicobacter pylori Resistance to Six Antibiotics by Two Breakpoint Systems and Resistance Evolution in Bulgaria. Infect. Dis. 2016, 48, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Jun, J.S.; Yeom, J.S.; Park, J.S.; Youn, H.S.; Ko, G.H.; Baik, S.C.; Lee, W.K.; Cho, M.J.; Rhee, K.H. Changing Pattern of Antibiotic Resistance of Helicobacter pylori in Children during 20 years in Jinju, South Korea. Pediatr. Int. 2013, 55, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Butenko, T.; Jeverica, S.; Orel, R.; Homan, M. Antibacterial Resistance and the Success of Tailored Triple Therapy in Helicobacter pylori Strains Isolated from Slovenian Children. Helicobacter 2017, 22, e12400. [Google Scholar] [CrossRef]
- Pastukh, N.; Peretz, A.; Brodsky, D.; Isakovich, N.; Azrad, M.; On, A. Antimicrobial Susceptibility of Helicobacter pylori Strains Isolated from Children in Israel. J. Glob. Antimicrob. Resist. 2018, 12, 175–178. [Google Scholar] [CrossRef]
- Megraud, F.; Coenen, S.; Versporten, A.; Kist, M.; Lopez-Brea, M.; Hirschl, A.M.; Andersen, L.P.; Goossens, H.; Glupczynski, Y.; Study Group Participants. Helicobacter pylori Resistance to Antibiotics in Europe and Its Relationship to Antibiotic Consumption. Gut 2013, 62, 34–42. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Mamishi, S.; Banar, M.; Keshavarz Valian, S.; Bahador, A.; Najafi, M.; Farahmand, F.; Pourakbari, B. Antibiotic Susceptibility of Helicobacter pylori Strains Isolated from Iranian Children: High Frequency of A2143G Point Mutation Associated with Clarithromycin Resistance. J. Glob. Antimicrob. Resist. 2017, 10, 131–135. [Google Scholar] [CrossRef]
- Güven, B.; Gülerman, F.; Kaçmaz, B. Helicobacter pylori Resistance to Clarithromycin and Fluoroquinolones in a Pediatric Population in Turkey: A Cross-Sectional Study. Helicobacter 2019, 24, e12581. [Google Scholar] [CrossRef]
- Iwańczak, B.M.; Borys-Iwanicka, A.; Biernat, M.; Gościniak, G. Assessment of Sequential and Standard Triple Therapy in Treatment of Helicobacter pylori Infection in Children Dependent on Bacteria Sensitivity to Antibiotics. Adv. Clin. Exp. Med. 2016, 25, 701–708. [Google Scholar] [CrossRef]
- Klesiewicz, K.; Nowak, P.; Karczewska, E.; Skiba, I.; Wojtas-Bonior, I.; Sito, E.; Budak, A. PCR-RFLP Detection of Point Mutations A2143G and A2142G in 23S rRNA Gene Conferring Resistance to Clarithromycin in Helicobacter pylori Strains. Acta Biochim. Pol. 2014, 61, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.; Burucoa, C.; Pietrini, O.; Bergeret, M.; Decoster, A.; Wann, A.; Dupont, C.; Kalach, N. Clarithromycin Resistance in Helicobacter pylori Strains Isolated from French Children: Prevalence of the Different Mutations and Coexistence of Clones Harboring Two Different Mutations in the Same Biopsy. Helicobacter 2007, 12, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wen, Y.; Xiao, Q.; Zheng, W.; Long, G.; Chen, B.; Shu, X.; Jiang, M. Mutations in the Antibiotic Target Genes Related to Clarithromycin, Metronidazole and Levofloxacin Resistance in Helicobacter pylori Strains from Children in China. Infect. Drug Resist. 2020, 13, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.I.; Do, B.J.; Kang, J.G.; Kim, H.S.; Jang, M.K.; Kim, H.Y.; Shin, W.G. Helicobacter pylori Eradication According to Sequencing-Based 23S Ribosomal RNA Point Mutation Associated with Clarithromycin Resistance. J. Clin. Med. 2019, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Pourakbari, B.; Mahmoudi, S.; Parhiz, J.; Sadeghi, R.H.; Monajemzadeh, M.; Mamishi, S. High Frequency of Metronidazole and Clarithromycin-Resistant Helicobacter pylori in Formalin-Fixed, Paraffin-Embedded Gastric Biopsies. Br. J. Biomed. Sci. 2018, 75, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, R.; Lionetti, E.; Castellaneta, S.; Margiotta, M.; Piscitelli, D.; Lorenzo, L.; Cavallo, L.; Ierardi, E. Clarithromycin-Resistant Genotypes and Eradication of Helicobacter pylori. J. Pediatr. 2010, 157, 228–232. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Y.; Lu, N.H. Novel and Effective Therapeutic Regimens for Helicobacter pylori in an Era of Increasing Antibiotic Resistance. Front. Cell. Infect. Microbiol. 2017, 7, 168. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori Biofilm Formation on Susceptibility to Amoxicillin, Metronidazole and Clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Binh, T.T.; Suzuki, R.; Trang, T.T.H.; Kwon, D.H.; Yamaoka, Y. Search for Novel Candidate Mutations for Metronidazole Resistance in Helicobacter pylori Using Next-Generation Sequencing. Antimicrob. Agents Chemother. 2015, 59, 2343–2348. [Google Scholar] [CrossRef]
- Marais, A.; Bilardi, C.; Cantet, F.; Mendz, G.L.; Mégraud, F. Characterization of the Genes rdxA and frxA Involved in Metronidazole Resistance in Helicobacter pylori. Res. Microbiol. 2003, 154, 137–144. [Google Scholar] [CrossRef]
- Tanih, N.F.; Ndip, L.M.; Ndip, R.N. Characterisation of the Genes Encoding Resistance to Metronidazole (rdxA and frxA) and Clarithromycin (the 23S-rRNA Genes) in South African Isolates of Helicobacter pylori. Ann. Trop. Med. Parasitol. 2011, 105, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Hulten, K.; Kato, M.; Kim, J.J.; Lee, M.; El-Zaatari, F.A.K.; Osato, M.S.; Graham, D.Y. DNA Sequence Analysis of rdxA and frxA from 12 Pairs of Metronidazole-Sensitive and -Resistant Clinical Helicobacter pylori Isolates. Antimicrob. Agents Chemother. 2001, 45, 2609–2615. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Chivers, P.T.; Berg, D.E. Point Mutations in Helicobacter pylori’s fur Regulatory Gene that Alter Resistance to Metronidazole, a Prodrug Activated by Chemical Reduction. PLoS ONE 2011, 6, e18236. [Google Scholar] [CrossRef] [PubMed][Green Version]
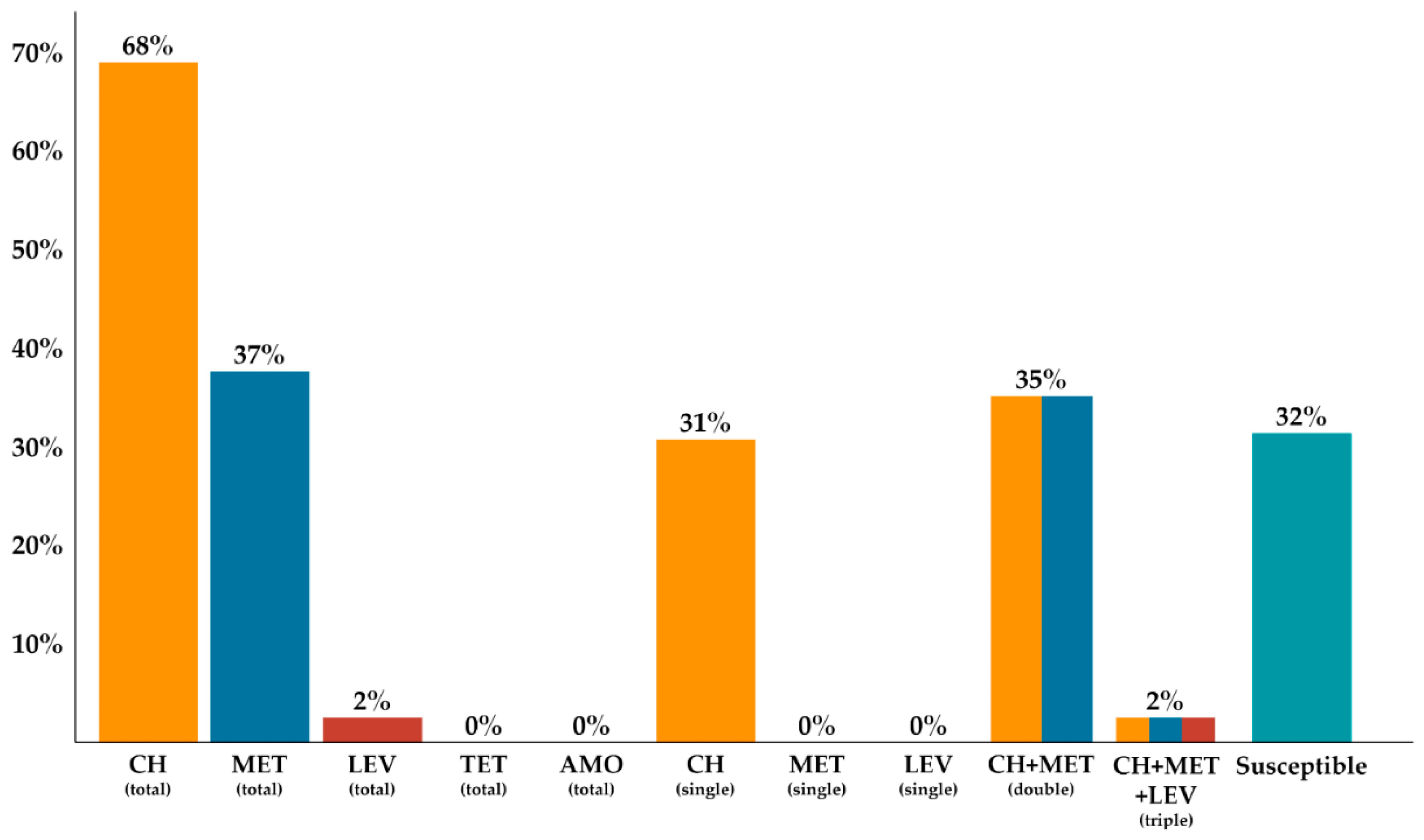
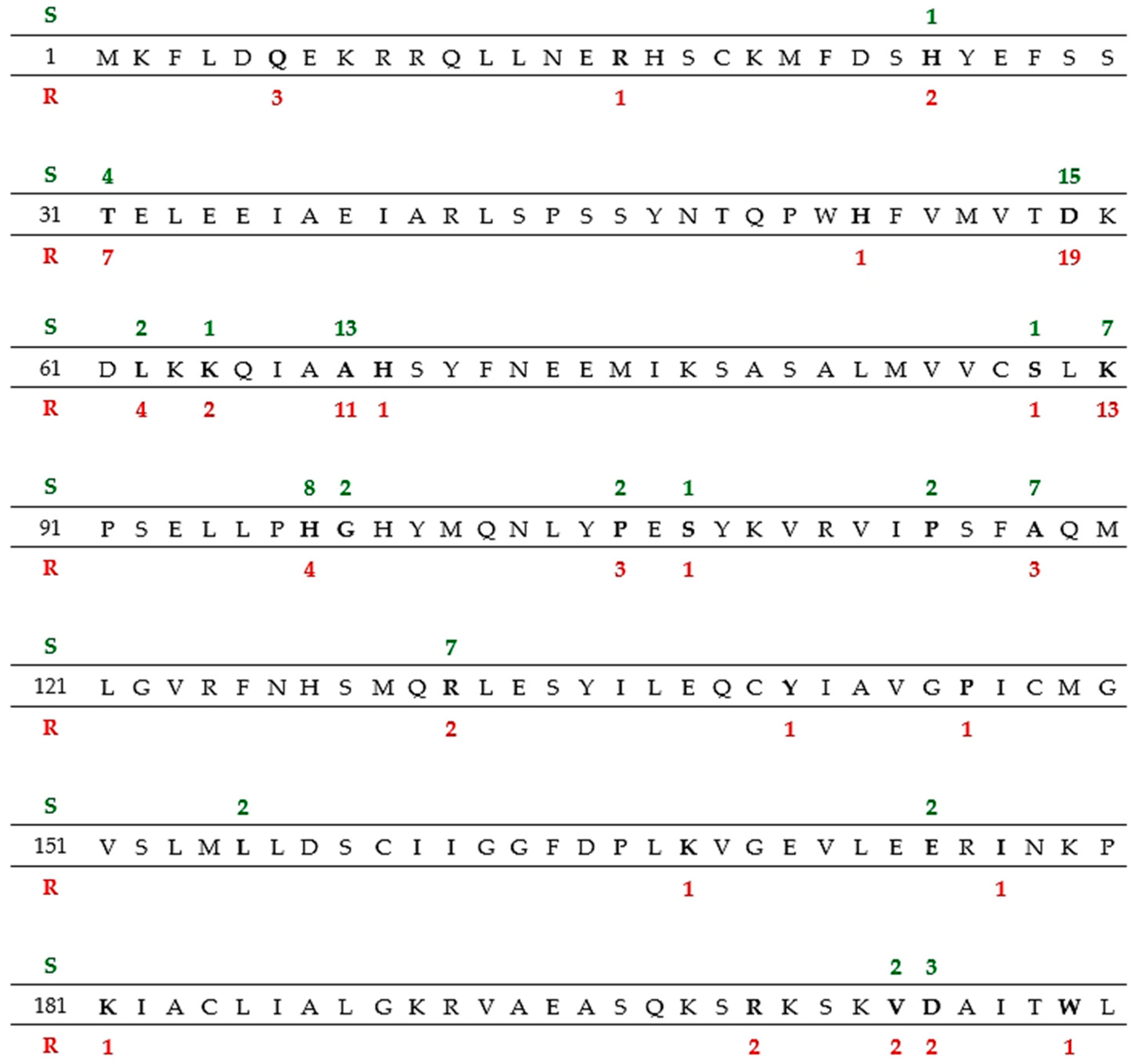
| H. pylori Strains | All Patients [Total] (n = 91) | Chronic Gastritis (n = 35) | GERD (n = 14) | Gastric/Duodenal Ulcer (n = 15) | Other * (n = 27) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | |
| Susceptible to CH | 17 (18.7%) | 12 (13.2%) | 7 (7.7%) | 5 (5.5%) | 3 (3.3%) | 1 (1.1%) | 1 (1.1%) | 0 (0.0%) | 4 (4.4%) | 8 (8.8%) |
| Resistant to CH | 25 (27.5%) | 37 (40.7%) | 13 (14.3%) | 10 (11%) | 4 (4.4%) | 6 (6.6%) | 6 (6.6%) | 8 (8.8%) | 5 (5.5%) | 10 (11%) |
| p = 0.032 | p > 0.05 | p = 0.01 | p = 0.03 | p > 0.05 | ||||||
| Susceptible to CH + MET | 17 (18.7%) | 40 (44%) | 7 (7.7%) | 13 (14.3%) | 3 (3.3%) | 7 (7.7%) | 1 (1.1%) | 8 (8.8%) | 4 (4.4%) | 18 (19.8%) |
| Resistant to CH + MET | 25 (27.5%) | 9 (9.9%) | 13 (14.3%) | 2 (2.2%) | 4 (4.4%) | 0 (0%) | 6 (6.6%) | 0 (0%) | 5 (5.5%) | 0 (0%) |
| p > 0.05 | p < 0.01 | p < 0.01 | p < 0.01 | p < 0.01 | ||||||
| Susceptible to CH + MET + LEV | 42 (46.1%) | 47 (51.6%) | 20 (22%) | 13 (14.3%) | 7 (7.7%) | 7 (7.7%) | 7 (7.7%) | 8 (8.8%) | 9 (9.9%) | 18 (19.8%) |
| Resistant to CH + MET + LEV | 0 (0%) | 2 (2.2%) | 0 (0%) | 2 (2.2%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| p < 0.01 | p < 0.01 | p > 0.05 | p > 0.05 | p > 0.05 | ||||||
| H. pylori Strains | Clinical Diagnosis | Significance | |||
|---|---|---|---|---|---|
| Chronic Gastritis (n = 35) | GERD (n = 14) | Peptic/Duodenal Ulcer (n = 15) | Other * (n = 27) | ||
| R/without mutation | 11% (4) | 7% (1) | 0% (0) | 4% (1) | p > 0.05 |
| R/A2142G | 8% (3) | 21% (3) | 7% (1) | 0% (0) | |
| R/A2143G | 40% (14) | 36% (5) | 80% (12) | 44% (12) | |
| R/T2182C | 6% (2) | 7% (1) | 7% (1) | 7% (2) | |
| S/without mutation | 23% (8) | 21% (3) | 7% (1) | 33% (9) | |
| S/A2143G | 3% (1) | 7% (1) | 0% (0) | 4% (1) | |
| S/T2182C | 9% (3) | 0% (0) | 0% (0) | 7% (2) | |
| H. pylori Strains | Number of Mutations | Number of Analyzed Positions without Mutations | Significance |
|---|---|---|---|
| MET-sensitive n = 2656 (16 × 166) | 387 | 2269 | p = 0.0361 (Chi2 = 4.3909) |
| MET-resistant n = 3154 (19 × 166) | 400 | 2754 |
| H. pylori Strains | All Patients | Mutations | Mutations per Patient | Stop Mutations | AA Changes n (%) | AA Changes per Patient |
|---|---|---|---|---|---|---|
| MET-sensitive | 16 | 387 | 24.2 | 1 | 82 (21%) | 5 |
| MET-resistant | 19 | 400 | 21 | 4 | 90 (22.5%) | 4.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biernat, M.M.; Bińkowska, A.; Łaczmański, Ł.; Biernat, P.; Krzyżek, P.; Gościniak, G. Phenotypic and Genotypic Analysis of Resistant Helicobacter pylori Strains Isolated from Children with Gastrointestinal Diseases. Diagnostics 2020, 10, 759. https://doi.org/10.3390/diagnostics10100759
Biernat MM, Bińkowska A, Łaczmański Ł, Biernat P, Krzyżek P, Gościniak G. Phenotypic and Genotypic Analysis of Resistant Helicobacter pylori Strains Isolated from Children with Gastrointestinal Diseases. Diagnostics. 2020; 10(10):759. https://doi.org/10.3390/diagnostics10100759
Chicago/Turabian StyleBiernat, Monika Maria, Aldona Bińkowska, Łukasz Łaczmański, Paweł Biernat, Paweł Krzyżek, and Grażyna Gościniak. 2020. "Phenotypic and Genotypic Analysis of Resistant Helicobacter pylori Strains Isolated from Children with Gastrointestinal Diseases" Diagnostics 10, no. 10: 759. https://doi.org/10.3390/diagnostics10100759
APA StyleBiernat, M. M., Bińkowska, A., Łaczmański, Ł., Biernat, P., Krzyżek, P., & Gościniak, G. (2020). Phenotypic and Genotypic Analysis of Resistant Helicobacter pylori Strains Isolated from Children with Gastrointestinal Diseases. Diagnostics, 10(10), 759. https://doi.org/10.3390/diagnostics10100759






