Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be?
Abstract
1. Introduction
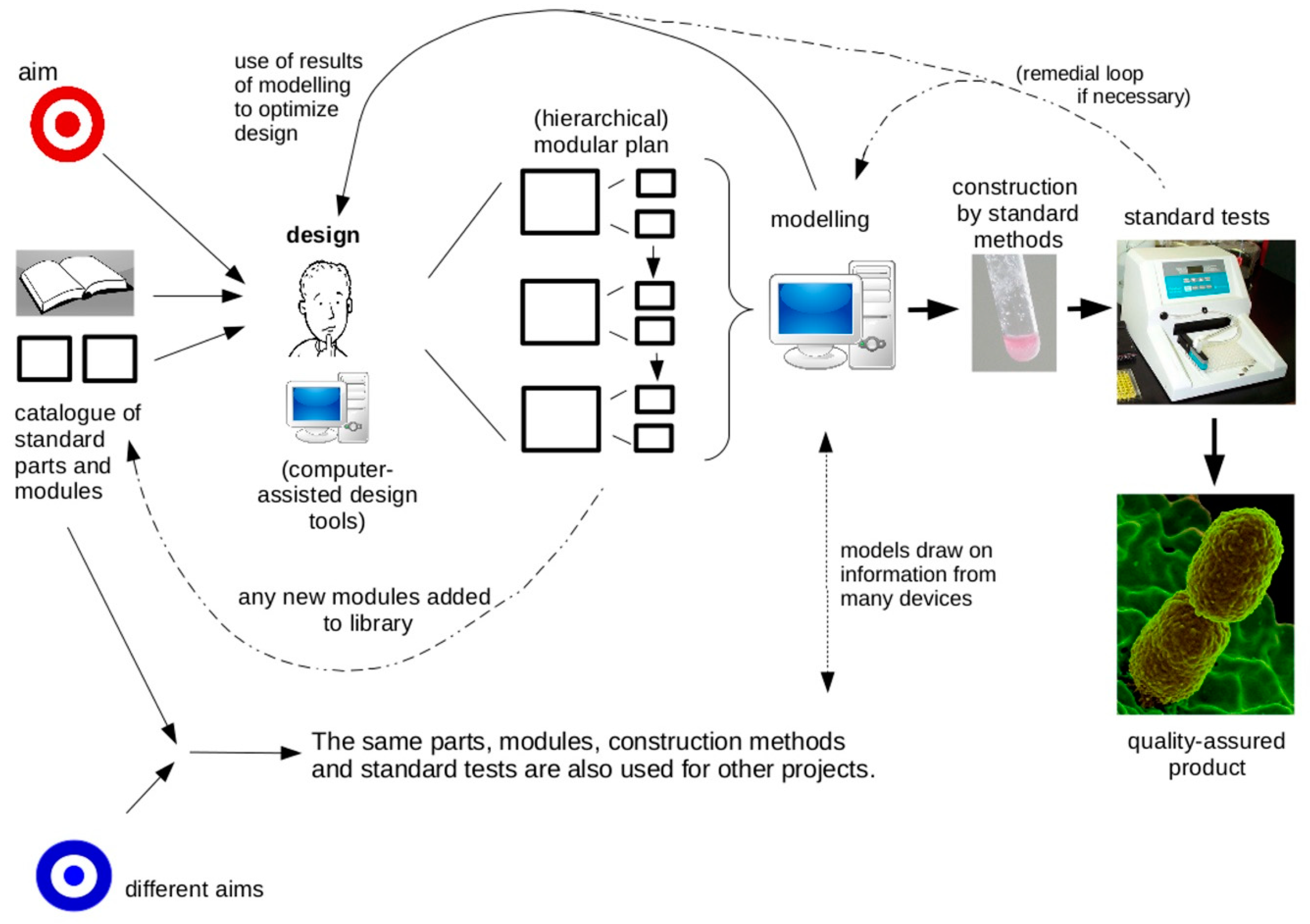
2. Features of the Engineering Approach
- a design phase that uses predictive models
- designing as much as possible using standard, well-characterised components
- hierarchical design using functional modules
- manufacture using reliable, quality-assured systems
- testing of (at least samples of) finished devices using standardised measurement techniques.
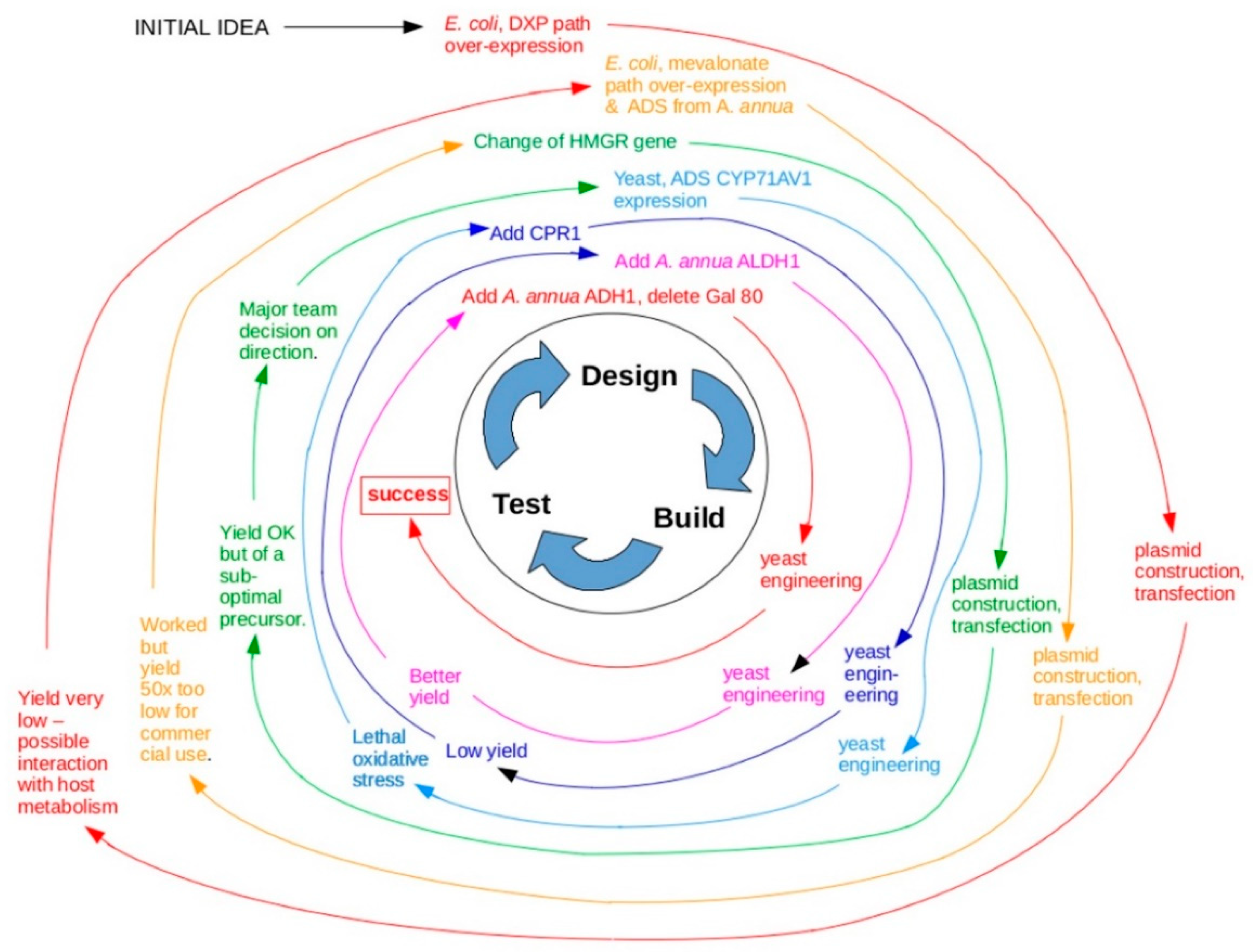
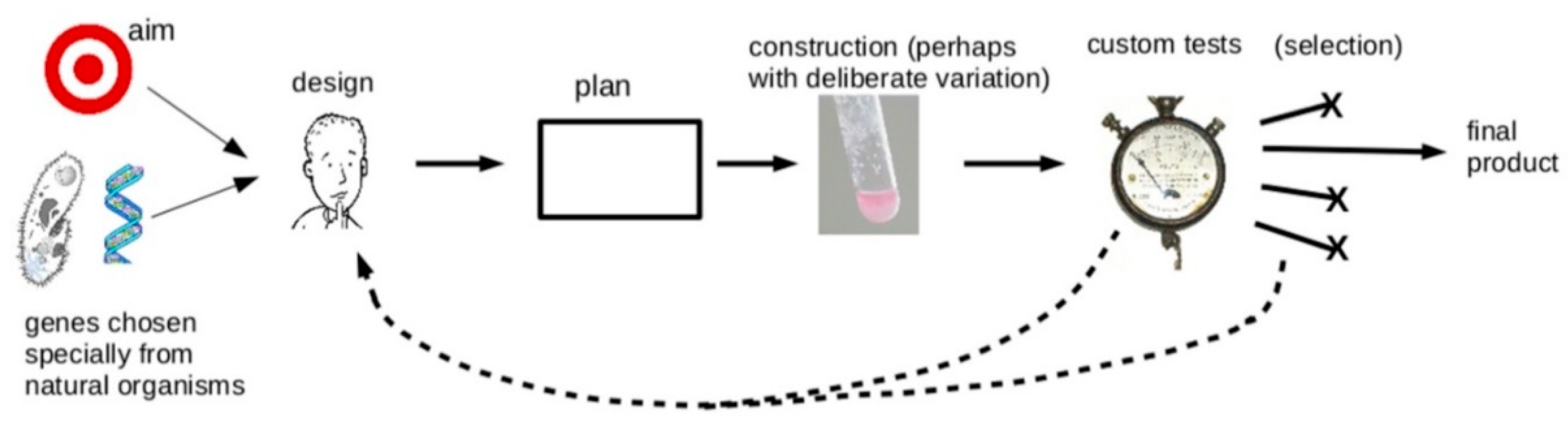
3. To What Extent Have Past Successes Followed the Engineering Approach?
4. Should Synthetic Biology Follow the Engineering Approach in the Future?
4.1. A Design Phase That Uses Predictive Models
4.2. Designing as Much as Possible Using Standard, Well-Characterized Components
4.3. Hierarchical Design Using Previously-Characterised Modules
4.4. Manufacture Using Reliable, Quality-Assured Systems
4.5. Testing of Devices Using Standardised Measurement Techniques
5. Where the Engineering Metaphor Is Useful
6. Conclusions
Funding
Conflicts of Interest
References
- Cheng, A.A.; Lu, T.K. Synthetic biology: An emerging engineering discipline. Annu. Rev. Biomed. Eng. 2012, 14, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.A. Synthetic Biology: A Very Short Introduction; Oxford University Press: Oxford, UK, 2018; pp. 3–9. [Google Scholar]
- Nielsen, J.; Keasling, J.D. Engineering Cellular Metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Jagadevan, S.; Banerjee, A.; Banerjee, C.; Guria, C.; Tiwari, R.; Baweja, M.; Shukla, P. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol. Biofuels 2018, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Cases, I.; de Lorenzo, V. Genetically modified organisms for the environment: Stories of success and failure and what we have learned from them. Int. Microbiol. 2005, 8, 213–222. [Google Scholar] [PubMed]
- Davies, J. Using synthetic biology to explore principles of development. Development 2017, 144, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Toparlak, O.D.; Mansy, S.S. Progress in synthesizing protocells. Exp. Biol. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Canton, B.; Labno, A.; Endy, D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008, 26, 787–793. [Google Scholar] [CrossRef]
- Fu, P. A perspective of synthetic biology: Assembling building blocks for novel functions. Biotechnol. J. 2006, 1, 690–699. [Google Scholar] [CrossRef]
- Royal Academy of Engineering. Synthetic Biology: Scope, Applications and Implications; Royal Academy of Engineering: London, UK, 2009. [Google Scholar]
- Shi, T.; Han, P.; You, C.; Zhang, Y.P.J. An in vitro synthetic biology platform for emerging industrial biomanufacturing: Bottom-up pathway design. Synth. Syst. Biotechnol. 2018, 3, 186–195. [Google Scholar] [CrossRef]
- Heinemann, M.; Panke, S. Synthetic biology-putting engineering into biology. Bioinformatics 2006, 22, 2790–2799. [Google Scholar] [CrossRef]
- Endy, D. Foundations for engineering biology. Nature 2005, 438, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Ghosh, S.; Kitano, H. Consistent design schematics for biological systems: Standardization of representation in biological engineering. J. R. Soc. Interface 2009, 6 (Suppl. 4), S393–S404. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Strelkowa, N.; Stan, G.-B.; Douglas, T.; Savulescu, J.; Barahona, M.; Papachristodoulou, A. Engineering and ethical perspectives in synthetic biology. EMBO Rep. 2012, 13, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Slusarczyk, A.L.; Lin, A.; Weiss, R. Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 2012, 13, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Hawkins, K. Synthetic biology: Evolution or revolution? A co-founder’s perspective. Curr. Opin. Chem. Biol. 2013, 17, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Kelwick, R.; Bowater, L.; Yeoman, K.H.; Bowater, R.P. Promoting microbiology education through the iGEM synthetic biology competition. FEMS Microbiol. Lett. 2015, 362, Fnv129. [Google Scholar] [CrossRef]
- Way, J.C.; Collins, J.J.; Keasling, J.D.; Silver, P.A. Integrating biological redesign: Where synthetic biology came from and where it needs to go. Cell 2014, 157, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.; Adams, J.; Bainbridge, J.; Birney, E.; Calvert, J.; Collis, A.; Kitney, R.; Freemont, P.; Mason, P.; Pandya, K.; et al. A Synthetic Biology Roadmap for the UK. Published by the UK Technology Strategy Board. 2012. Available online: https://connect.innovateuk.org/documents/2826135/3815409/Synthetic+Biology+Roadmap+-+Report.pdf/fa8a1e8e-cbf4-4464-87ce-b3b033f04eaa (accessed on 20 November 2018).
- Clarke, L.; Freeman, G.; Bainbridge, J.; Collis, A.; Dafforn, T.; Dunkerton, S.; Fell, T.; Jones, C.; Kent, A.; Kitney, R.; et al. Biodesign for the Bioeconomy: UK Synthetic Biology Strategic Plan 2016. Published by the Synthetic Biology Leadership Council. 2016. Available online: https://connect.innovateuk.org/documents/2826135/31405930/BioDesign+for+the+Bioeconomy+2016+DIGITAL+updated+21_03_2016.pdf/d0409f15-bad3-4f55-be03-430bc7ab4e7e (accessed on 20 November 2018).
- Tong, S.; Zhao, H. A brief overview of synthetic biology research programs and roadmap studies in the United States. Synth. Syst. Biol. 2016, 1, 258–264. [Google Scholar]
- Shih, P.M. Towards a sustainable bio-based economy: Redirecting primary metabolism to new products with plant synthetic biology. Plant Sci. 2018, 273, 84–91. [Google Scholar] [CrossRef]
- French, C.E. Synthetic biology and biomass conversion: A match made in heaven? J. R. Soc. Interface 2009, 6 (Suppl. 4), S547–S558. [Google Scholar] [CrossRef]
- Goold, H.D.; Wright, P.; Hailstones, D. Emerging Opportunities for Synthetic Biology in Agriculture. Genes (Basel) 2018, 9, E341. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.S.; Collins, J.J. Synthetic biology: Applications come of age. Nat. Rev. Genet. 2010, 11, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Synbioproject Applications Database. Available online: http://synbioproject.org/cpi/applications/ (accessed on 16 December 2018).
- Kwok, R. Five hard truths for synthetic biology. Nature 2010, 463, 288–290. [Google Scholar] [CrossRef]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Genencor Product Information Sheet for Accelerase Trio. Available online: http://www.genencor.com/fileadmin/user_upload/genencor/documents/TRIO_ProductSheet_LowRes.pdf (accessed on 6 December 2018).
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Keasling, J.D. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014, 12, 355–367. [Google Scholar] [CrossRef] [PubMed]
- ETC Group. Artemisinin and Synthetic Biology: A Case Study. 2014. Available online: http://www.etcgroup.org/sites/www.etcgroup.org/files/ETC-artemisinin-synbio-casestudy2014.pdf (accessed on 20 November 2018).
- Kim, J.H.; Wang, C.; Jang, H.J.; Cha, M.S.; Park, J.E.; Jo, S.Y.; Choi, E.S.; Kim, S.W. Isoprene production by Escherichia coli through the exogenous mevalonate pathway with reduced formation of fermentation byproducts. Microb. Cell Fact. 2016, 15, 214. [Google Scholar] [CrossRef]
- Novozyme Product Information Sheet for Cellic CTec. Available online: http://www.shinshu-u.ac.jp/faculty/engineering/chair/chem010/manual/Ctec2.pdf (accessed on 6 December 2018).
- Müller, U.; van Assema, F.; Gunsior, M.; Orf, S.; Kremer, S.; Schipper, D.; Wagemans, A.; Townsend, C.A.; Sonke, T.; Bovenberg, R.; et al. Metabolic engineering of the E. coli L-phenylalanine pathway for the production of D-phenylglycine (D-Phg). Metab. Eng. 2006, 8, 196–208. [Google Scholar] [CrossRef]
- Zhang, F.; Carothers, J.M.; Keasling, J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012, 30, 354–359. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, P.; Zhang, X.; Dong, M. Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2017, 44, 1083–1095. [Google Scholar] [CrossRef]
- de Mora, K.; Joshi, N.; Balint, B.L.; Ward, F.B.; Elfick, A.; French, C.E. A pH-based biosensor for detection of arsenic in drinking water. Anal. Bioanal. Chem. 2011, 400, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, J.; Bizzari, F.; Cai, Y.; Davidson, B.; de Mora, K.; Ivakhno, S.; Seshasayee, S.L.; Nicholson, J.; Wilson, J.; Elfick, A.; et al. Development of a novel biosensor for the detection of arsenic in drinking water. IET Synth. Biol. 2007, 1, 87–90. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Oakes, B.L.; Nadler, D.C.; Flamholz, A.; Fellmann, C.; Staahl, B.T.; Doudna, J.A.; Savage, D.F. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol. 2016, 34, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Monedero, A.; Davies, J.A. Tamoxifen- and Mifepristone-Inducible Versions of CRISPR Effectors, Cas9 and Cpf1. ACS Synth. Biol. 2018, 7, 2160–2169. [Google Scholar] [CrossRef] [PubMed]
- Decoene, T.; De Paepe, B.; Maertens, J.; Coussement, P.; Peters, G.; De Maeseneire, S.L.; De Mey, M. Standardization in synthetic biology: An engineering discipline coming of age. Crit. Rev. Biotechnol. 2018, 38, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Hammond, R. Building the Business of Biodesign: The Synthetic Biology Industry is Ready to Change Gear. Cambridge Consultants Workshop Report. 2018. Available online: https://www.cambridgeconsultants.com/sites/default/files/uploaded-pdfs/Building%20the%20business%20of%20biodesign%20%28workshop%20report%29_0.pdf (accessed on 20 November 2018).
- Müller, K.M.; Arndt, K.M. Standardization in synthetic biology. Methods Mol. Biol. 2012, 813, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Appleton, E.; Madsen, C.; Roehner, N.; Densmore, D. Design Automation in Synthetic Biology. Cold Spring Harb. Perspect. Biol. 2017, 9, A023978. [Google Scholar] [CrossRef]
- Peccoud, J.; Blauvelt, M.F.; Cai, Y.; Cooper, K.L.; Crasta, O.; DeLalla, E.C.; Evans, C.; Folkerts, O.; Lyons, B.M.; Mane, S.P.; et al. Targeted development of registries of biological parts. PLoS ONE 2008, 3, e2671. [Google Scholar] [CrossRef]
- Whitworth, J. A Paper on an Uniform System of Screw Threads. Read at the Institute of Civil Engineers. 1841. Available online: https://en.wikisource.org/wiki/Miscellaneous_Papers_on_Mechanical_Subjects/A_Paper_on_an_Uniform_System_of_Screw_Threads (accessed on 20 November 2018).
- Knight, T. Idempotent Vector Design for Standard Assembly of Biobricks. MIT Artificial Intelligence Laboratory. 2003. Available online: http://hdl.handle.net/1721.1/21168 (accessed on 20 November 2018).
- Knight, T. Draft Standard for Biobrick Biological Parts; OpenWetWare, MIT: Cambridge, MA, USA, 2007; Available online: http://hdl.handle.net/1721.1/45138 (accessed on 18 November 2018).
- Martin, M.D.; Cappellini, E.; Samaniego, J.A.; Zepeda, M.L.; Campos, P.F.; Seguin-Orlando, A.; Wales, N.; Orlando, L.; Ho, S.Y.; Dietrich, F.S.; et al. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 2013, 4, 2172. [Google Scholar] [CrossRef]
- Scales, J. What Happened When I Peeked into My NODE_Modules Directory: Just Absolute Madness. 2016. Available online: https://medium.com/s/silicon-satire/i-peeked-into-my-node-modules-directory-and-you-wont-believe-what-happened-next-b89f63d21558 (accessed on 20 November 2018).
- Propokov, N. Software Disenchantment. 2018. Available online: http://tonsky.me/blog/disenchantment/ (accessed on 18 November 2018).
- Pedersen, M.; Phillips, A. Towards programming languages for genetic engineering of living cells. J. R. Soc. Interface 2009, 6 (Suppl. 4), S437–S450. [Google Scholar] [CrossRef] [PubMed]
- Greber, D.; Fussenegger, M. Mammalian synthetic biology: Engineering of sophisticated gene networks. J. Biotechnol. 2007, 130, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Kittleson, J.T.; Wu, G.C.; Anderson, J.C. Successes and failures in modular genetic engineering. Curr. Opin. Chem. Biol. 2012, 16, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Sainz de Murieta, I.; Bultelle, M.; Kitney, R.I. Toward the First Data Acquisition Standard in Synthetic Biology. ACS Synth. Biol. 2016, 5, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C. Engineering ingenuity at iGEM. Nat. Chem. Biol. 2008, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Røkke, G.; Korvald, E.; Pahr, J.; Oyås, O.; Lale, R. BioBrick assembly standards and techniques and associated software tools. Methods Mol. Biol. 2014, 1116, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, C.; Porcar, M. iGEM 2.0-refoundations for engineering biology. Nat. Biotechnol. 2014, 32, 420–424. [Google Scholar] [CrossRef]
- Joshi, N.; Wang, X.; Montgomery, L.; Elfick, A.; French, C.E. Novel approaches to biosensors for detection of arsenic in drinking water. Desalination 2009, 248, 517–523. [Google Scholar] [CrossRef]
- French, C.E.; de Mora, K.; Joshi, N.; Elfick, A.; Haseloff, J.; Ajioka, J. Synthetic biology and the art of biosensor design. In the Science and Applications of Synthetic and Systems Biology: Workshop Summary; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Arsenic Biosensor Collabroation. Can We Build a Cheap, Practical and Reliable Arsenic Biosensor? 2014. Available online: http://www.arsenicbiosensor.org/team.html (accessed on 15 November 2018).
- Brazma, A.; Hingamp, P.; Quackenbush, J.; Sherlock, G.; Spellman, P.; Stoeckert, C.; Aach, J.; Ansorge, W.; Ball, C.A.; Causton, H.C.; et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001, 29, 365–371. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Ball, C.A.; Bova, G.S.; Brazma, A.; Bumgarner, R.E.; Campbell, D.; Causton, H.C.; Christiansen, J.; Davidson, D.; Eichner, L.J.; et al. Development of the Minimum Information Specification for In Situ Hybridization and Immunohistochemistry Experiments (MISFISHIE). OMICS 2006, 10, 205–208. [Google Scholar] [CrossRef]
- Taylor, C.F.; Field, D.; Sansone, S.A.; Aerts, J.; Apweiler, R.; Ashburner, M.; Ball, C.A.; Binz, P.A.; Bogue, M.; Booth, T.; et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: The MIBBI project. Nat. Biotechnol. 2008, 26, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Robles, G.; González-Barahona, J.M. A Comprehensive Study of Software Forks: Dates, Reasons and Outcomes. In IFIP Advances in Information and Communication Technology; Hammouda, I., Lundell, B., Mikkonen, T., Scacchi, W., Eds.; Open Source Systems: Long-Term Sustainability OSS; Springer: Berlin/Heidelberg, Germany, 2012; Volume 378. [Google Scholar]
- Enna, S.J. Phenotypic drug screening. J. Peripher. Nerv. Syst. 2014, 19 (Suppl. 2), S4–S5. [Google Scholar] [CrossRef]
- Paricharak, S.; IJzerman, A.P.; Bender, A.; Nigsch, F. Analysis of Iterative Screening with Stepwise Compound Selection Based on Novartis In-house HTS Data. ACS Chem. Biol. 2016, 11, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, F.; Medina, D.L.; De Leo, E.; Panarella, A.; Emma, F. High-content drug screening for rare diseases. J. Inherit. Metab. Dis. 2017, 40, 601–607. [Google Scholar] [CrossRef]
- Gilbert, N. Cross-bred crops get fit faster. Nature 2014, 513, 292. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.H. Adaptation in Natural and Artificial Systems; MIT Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Poon, K.F.; Conway, G.; Wardrop, G.; Mellis, J. Successful Application of Genetic Algorithms to Network Design and Planning. BT Technol. J. 2000, 18, 32–41. [Google Scholar] [CrossRef]
- Hornby, G.; Globus, A.; Linden, D.S.; Lohn, J.D. Automated Antenna Design with Evolutionary Algorithms. White Paper Published by American Institute of Aeronatics and Astronautics. 2006. Available online: http://alglobus.net/NASAwork/papers/Space2006Antenna.pdf (accessed on 20 November 2018).
| Project | How | Ref | Status | Predictive Models? | Std Components? | Modular? | Standard Testing? |
|---|---|---|---|---|---|---|---|
| Accelerase Trio (for biomass conversion) | Enzyme engineering | [30] | Commercial | Unknown | No (started with natural enzymes chosen for this specific purpose) | No | No (custom) |
| Arteminisic acid synthesis (drug) | Metabolic path engineering | [31,32,33] | Commercial | No (design was optimized empirically) | No (custom, selected from other organisms) | No | No (custom) |
| Bioisoprene synthesis (for rubber) | Metabolic path engineering | [34] | Near-market | No (empirical testing of alternatives was used instead) | No (custom, selected from other organisms) | No | No (custom) |
| Cellic Ctec (enzyme for biomass conversion) | Enzyme engineering | [35] | Commercial | Unknown | Enzyme engineering | No | No (custom) |
| Cephalexin synthesis (drug) | Metabolic path engineering | [36] | Commercial | No (design was optimized empirically) | No (custom, selected from other organisms) | No | No (custom) |
| Biodiesel | Metabolic path engineering | [37] | Near-market | No (a model was used to inform overall plan and to assist with analysis but not to predict based on components) | No (custom, selected from other organisms) | No | No (custom) |
| Fuelzyme amylase | Enzyme engineering | [27] | Commerical | Unknown | No (custom, selected from other organisms) | No | No (custom) |
| Luminase PB-100 | Enzyme engineering | [27] | Commercial | Unknown | No (custom) | No | No (custom) |
| Resveratrol synthesis (drug) | Metabolic path engineering | [38] | Near-market | No (design was optimized empirically) | No (custom, selected from other organisms) | No | No (custom) |
| Arsenic sensing | Novel genetic ‘circuit’ with sensor | [39,40] | Near-market | Yes (ODE-based model using estimated parameters for performance prediction and sensitivity analysis) | Yes (BioBricks) | No | Partially |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davies, J.A. Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be? Life 2019, 9, 6. https://doi.org/10.3390/life9010006
Davies JA. Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be? Life. 2019; 9(1):6. https://doi.org/10.3390/life9010006
Chicago/Turabian StyleDavies, Jamie A. 2019. "Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be?" Life 9, no. 1: 6. https://doi.org/10.3390/life9010006
APA StyleDavies, J. A. (2019). Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be? Life, 9(1), 6. https://doi.org/10.3390/life9010006





