Regeneration of Escherichia coli Giant Protoplasts to Their Original Form
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents and Bacterial Strains
2.2. Giant Protoplast Preparation
2.3. Recovery from GP
2.4. Microchamber Array Fabrication
2.5. Microscopic Observations and Analysis
3. Results
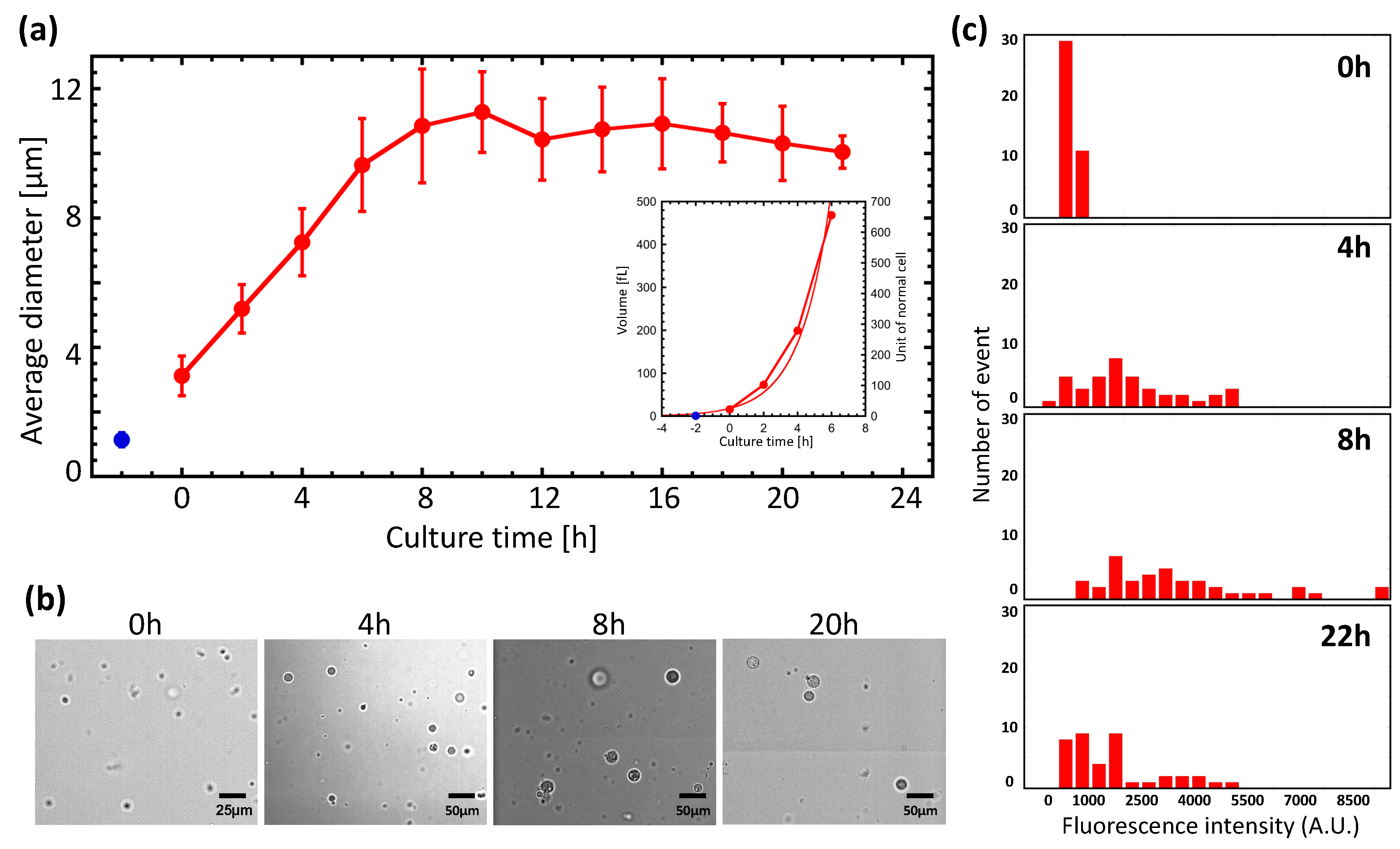
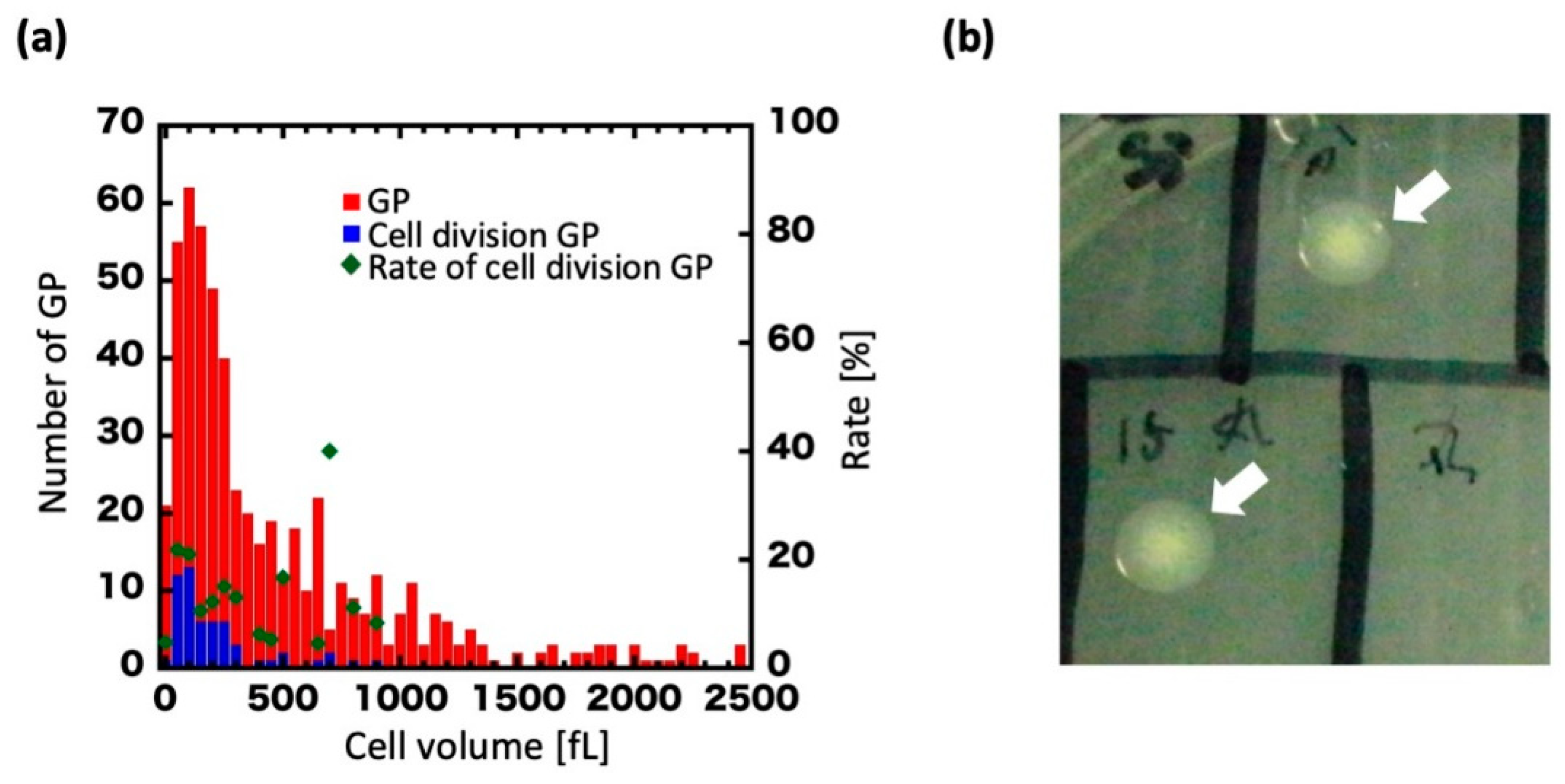
3.1. Giant Protoplast (GP) Size and Incubation Time
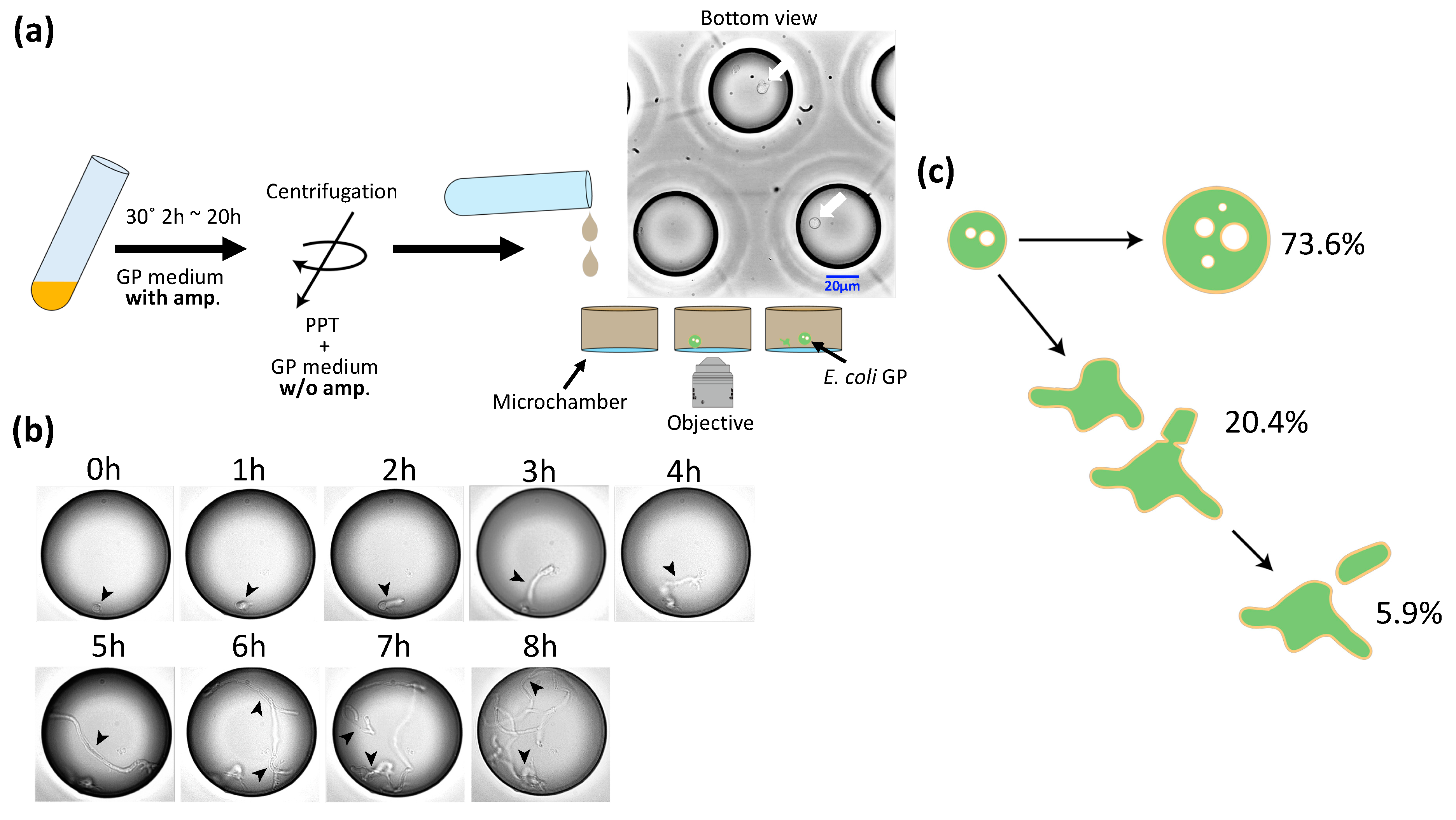
3.2. Regeneration of GPs
3.3. Deformation and Division of GPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Horinouchi, T.; Tamaoka, K.; Furusawa, C.; Ono, N.; Suzuki, S.; Hirasawa, T.; Yomo, T.; Shimizu, H. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genom. 2010, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Yomano, L.P.; York, S.W.; Ingram, L.O. Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J. Ind. Microbiol. Biotechnol. 1998, 20, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.W.; Matsumoto, Y.; Kitahara, K.; Suzuki, S.; Ono, N.; Furusawa, C.; Kishimoto, T.; Yomo, T. Bacterial transcriptome reorganization in thermal adaptive evolution. BMC Genom. 2015, 16, 802. [Google Scholar] [CrossRef] [PubMed]
- Barcenas-Moreno, G.; Gomez-Brandon, M.; Rousk, J.; Baath, E. Adaptation of soil microbial communities to temperature: Comparison of fungi and bacteria in a laboratory experiment. Glob. Chang. Biol. 2009, 15, 2950–2957. [Google Scholar] [CrossRef]
- Leroi, A.M.; Lenski, R.E.; Bennett, A.F. Evolutionary Adaptation to Temperature. III. Adaptation of Escherichia coli to a Temporally Varying Environment. Evolution 1994, 48, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Wakamoto, Y.; Dhar, N.; Chait, R.; Schneider, K.; Signorino-Gelo, F.; Leibler, S.; McKinney, J.D. Dynamic persistence of antibiotic-stressed mycobacteria. Science 2013, 339, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Moriizumi, Y.; Tabata, K.V.; Watanabe, R.; Doura, T.; Kamiya, M.; Urano, Y.; Noji, H. Hybrid cell reactor system from Escherichia coli protoplast cells and arrayed lipid bilayer chamber device. Sci. Rep. 2018, 8, 11757. [Google Scholar] [CrossRef] [PubMed]
- Fodor, K.; Alfoldi, L. Fusion of protoplasts of Bacillus megaterium. Proc. Natl. Acad. Sci. USA 1976, 73, 2147–2150. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.L. Protoplast formation in Escherichia coli. J. Bacteriol. 1976, 128, 668–670. [Google Scholar] [PubMed]
- Beran, V.; Havelkova, M.; Kaustova, J.; Dvorska, L.; Pavlik, I. Cell wall deficient forms of mycobacteria: A review. Vet. Med. 2006, 51, 365–389. [Google Scholar] [CrossRef]
- Mattman, L.H. Cell Wall Deficient Forms: Stealth Pathogens, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2000. [Google Scholar]
- Martin, H.H. Bacterial Protoplasts—A Review. J. Theor. Biol. 1963, 5, 1–34. [Google Scholar] [CrossRef]
- Mohan, R.R.; Kronish, D.P.; Pianotti, R.S.; Epstein, R.L.; Schwartz, B.S. Autolytic Mechanism for Spheroplast Formation in Bacillus cereus and Escherichia coli. J. Bacteriol. 1965, 90, 1355–1364. [Google Scholar] [PubMed]
- Salton, M.R.J. Cell Wall of Micrococcus-Lysodeikticus as the Substrate of Lysozyme. Nature 1952, 170, 746–747. [Google Scholar] [CrossRef] [PubMed]
- Weibull, C. The Isolation of Protoplasts from Bacillus megaterium by Controlled Treatment with Lysozyme. J. Bacteriol. 1953, 66, 688–695. [Google Scholar] [PubMed]
- Abrams, A. Reversible metabolic swelling of bacterial protoplasts. J. Biol. Chem. 1959, 234, 383–388. [Google Scholar] [PubMed]
- Galbraith, H.; Miller, T.B. Effect of long chain fatty acids on bacterial respiration and amino acid uptake. J. Appl. Bacteriol. 1973, 36, 659–675. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Ziesman, S.; Ratcliffe, T.; Gill, R.T.; Copley, S.D. Visualization of protoplast fusion and quantitation of recombination in fused protoplasts of auxotrophic strains of Escherichia coli. Metab. Eng. 2005, 7, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Fukui, K.; Sagara, Y.; Yoshida, N.; Matsuoka, T. Analytical studies on regeneration of protoplasts of Geotrichum candidum by quantitative thin-layer-agar plating. J. Bacteriol. 1969, 98, 256–263. [Google Scholar] [PubMed]
- Hopwood, D.A. Genetic studies with bacterial protoplasts. Annu. Rev. Microbiol. 1981, 35, 237–272. [Google Scholar] [CrossRef] [PubMed]
- Hadlaczky, G.; Fodor, K.; Alfoldi, L. Morphological study of the reversion to bacillary form, of Bacillus megaterium protoplasts. J. Bacteriol. 1976, 125, 1172–1179. [Google Scholar] [PubMed]
- Kubalski, A. Generation of giant protoplasts of Escherichia coli and an inner-membrane anion selective conductance. Biochim. Biophys. Acta 1995, 1238, 177–182. [Google Scholar] [CrossRef]
- Kusaka, I. Growth and division of protoplasts of Bacillus megaterium and inhibition of division by penicillin. J. Bacteriol. 1967, 94, 884–888. [Google Scholar] [PubMed]
- Lederberg, J.; St Clair, J. Protoplasts and L-type growth of Escherichia coli. J. Bacteriol. 1958, 75, 143–160. [Google Scholar] [PubMed]
- Churchward, G.; Estiva, E.; Bremer, H. Growth rate-dependent control of chromosome replication initiation in Escherichia coli. J. Bacteriol. 1981, 145, 1232–1238. [Google Scholar] [PubMed]
- Fagerbakke, K.M.; Heldal, M.; Norland, S. Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat. Microb. Ecol. 1996, 10, 15–27. [Google Scholar] [CrossRef]
- Kubitschek, H.E.; Friske, J.A. Determination of bacterial cell volume with the Coulter Counter. J. Bacteriol. 1986, 168, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Loferer-Krossbacher, M.; Klima, J.; Psenner, R. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 1998, 64, 688–694. [Google Scholar] [PubMed]
- Sullivan, C.J.; Morrell, J.L.; Allison, D.P.; Doktycz, M.J. Mounting of Escherichia coli spheroplasts for AFM imaging. Ultramicroscopy 2005, 105, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Ikeda, S.; Matsuo, T.; Hirata, A.; Takehara, M.; Hiyama, T.; Kawamura, F.; Kusaka, I.; Tsuchiya, T.; Kuroda, T.; et al. Patch clamp analysis of the respiratory chain in Bacillus subtilis. Biochim. Biophys. Acta 2011, 1808, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, T.; Okuda, N.; Saitoh, N.; Hiyama, T.; Terasaki, Y.; Anazawa, H.; Hirata, A.; Mogi, T.; Kusaka, I.; Tsuchiya, T.; et al. Patch clamp studies on ion pumps of the cytoplasmic membrane of Escherichia coli. Formation, preparation, and utilization of giant vacuole-like structures consisting of everted cytoplasmic membrane. J. Biol. Chem. 1998, 273, 16897–16904. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of contractile FtsZ rings in liposomes. Science 2008, 320, 792–794. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Kochanowski, K.; Vedelaar, S.; Ahrne, E.; Volkmer, B.; Callipo, L.; Knoops, K.; Bauer, M.; Aebersold, R.; Heinemann, M. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016, 34, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, T.; Sekiguchi, J. Studies on Regeneration Media for Bacillus-subtilis Protoplasts. Agric. Biol. Chem. 1981, 45, 2887–2894. [Google Scholar] [CrossRef]
- Glover, W.A.; Yang, Y.; Zhang, Y. Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS ONE 2009, 4, e7316. [Google Scholar] [CrossRef] [PubMed]
- Allan, E.; Hoischen, C.; Gumpert, J. Bacterial L-Forms. Adv. Appl. Microbiol. 2009, 68, 1–39. [Google Scholar] [PubMed]
- Onoda, T.; Enokizono, J.; Kaya, H.; Oshima, A.; Freestone, P.; Norris, V. Effects of calcium and calcium chelators on growth and morphology of Escherichia coli L-form NC-7. J. Bacteriol. 2000, 182, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- Katz, W.; Martin, H.H. Peptide crosslinkage in cell wall murein of Proteus mirabilis and its penicillin-induced unstable L-form. Biochem. Biophys. Res. Commun. 1970, 39, 744–749. [Google Scholar] [CrossRef]
- Leaver, M.; Domínguez-Cuevas, P.; Coxhead, J.M.; Daniel, R.A.; Errington, J. Life without a wall or division machine in Bacillus subtilis. Nature 2009, 457, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Errington, J. L-form bacteria, cell walls and the origins of life. Open Biol. 2013, 3, 120143. [Google Scholar] [CrossRef] [PubMed]
- Errington, J.; Mickiewicz, K.; Kawai, Y.; Wu, L.J. L-form bacteria, chronic diseases and the origins of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef] [PubMed]
- Joseleau-Petit, D.; Liébart, J.-C.; Ayala, J.A.; D’Ari, R. Unstable Escherichia coli L. forms revisited: Growth requires peptidoglycan synthesis. J. Bacteriol. 2007, 189, 6512–6520. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; Kawai, Y.; Errington, J. Wall proficient E. coli capable of sustained growth in the absence of the Z-ring division machine. Nat. Microbiol. 2016, 1, 16091. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabata, K.V.; Sogo, T.; Moriizumi, Y.; Noji, H. Regeneration of Escherichia coli Giant Protoplasts to Their Original Form. Life 2019, 9, 24. https://doi.org/10.3390/life9010024
Tabata KV, Sogo T, Moriizumi Y, Noji H. Regeneration of Escherichia coli Giant Protoplasts to Their Original Form. Life. 2019; 9(1):24. https://doi.org/10.3390/life9010024
Chicago/Turabian StyleTabata, Kazuhito V., Takao Sogo, Yoshiki Moriizumi, and Hiroyuki Noji. 2019. "Regeneration of Escherichia coli Giant Protoplasts to Their Original Form" Life 9, no. 1: 24. https://doi.org/10.3390/life9010024
APA StyleTabata, K. V., Sogo, T., Moriizumi, Y., & Noji, H. (2019). Regeneration of Escherichia coli Giant Protoplasts to Their Original Form. Life, 9(1), 24. https://doi.org/10.3390/life9010024




