Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Isolation
2.2. DNA Extraction, PCR Amplification, and DNA Sequencing
2.3. Alignment and Tree Reconstruction
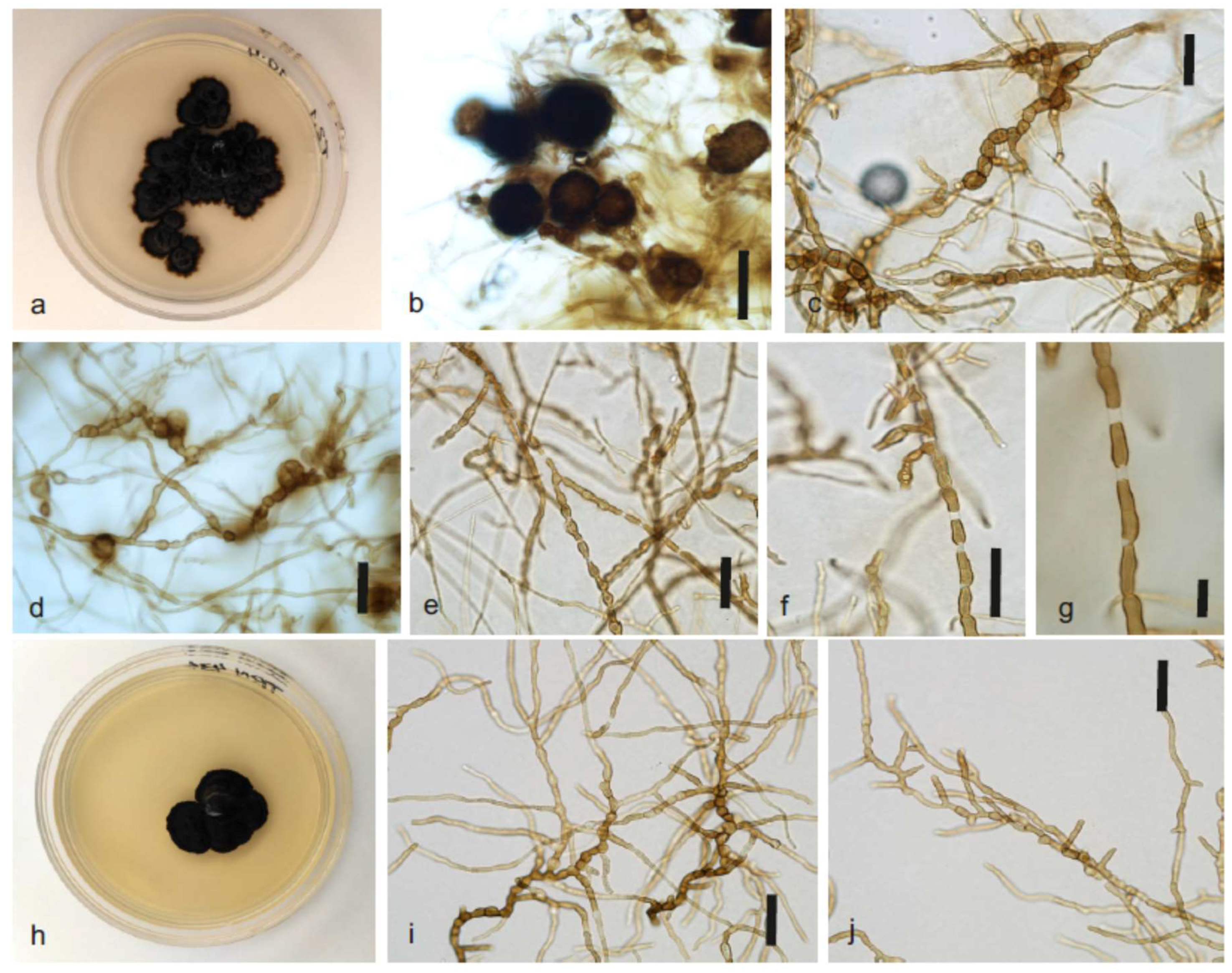
2.4. Morphological Characterization
3. Results
3.1. Subaerial Biofilm Development and Isolation
3.2. Strains Analyzed
3.3. Sequences and BLAST Search
3.4. Alignment and Tree Reconstruction
3.5. Taxonomy of Constantinomyces oldenburgensis sp. nov. and Constantinomyces patonensis sp. nov.
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gorbushina, A.A. Life on the rocks. Environ. Microbiol. 2007, 9, 1613–1631. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M. Fungi, rocks, and minerals. Elements 2017, 13, 171–176. [Google Scholar] [CrossRef]
- Staley, J.T.; Palmer, F.; Adams, J.B. Microcolonial fungi: Common inhabitants on desert rocks? Science 1982, 215, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Urzì, C.; Krumbein, W.E.; Warscheid, T. On the question of biogenic color changes of Mediterranean monuments (Coatings, Crust, Microstromatolite, Patina, Scialbatura, Skin, Rock Varnish). In Proceedings of the 2nd International Symposium “The Conservation of Monuments in the Mediterranean Basin, Genève, Switzerland, 19–21 November 1991. [Google Scholar]
- Wollenzien, U.; De Hoog, G.S.; Krumbein, W.E.; Urzi, C. On the isolation of microcolonial fungi occurring on and in marble and other calcareous rocks. Sci. Total Environ. 1995, 167, 287–294. [Google Scholar] [CrossRef]
- Sterflinger, K.; Krumbein, W.E. Dematiaceous fungi as a major agent for biopitting on Mediterranean marbles and limestones. Geomicrobiol. J. 1997, 14, 219–230. [Google Scholar] [CrossRef]
- Ruibal, C.; Platas, G.; Bills, G.F. Isolation and characterization of melanized fungi from limestone formations in Mallorca. Mycol. Prog. 2005, 4, 23–38. [Google Scholar] [CrossRef]
- Ruibal, C.; Platas, G.; Bills, G.F. High diversity and morphological convergence among melanized fungi from rock formations in the Central Mountain System of Spain. Persoonia 2008, 21, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Isola, D.; Zucconi, L.; Onofri, S.; Caneva, G.; De Hoog, G.S.; Selbmann, L. Extremotolerant rock inhabiting black fungi from Italian monumental sites. Fungal Divers. 2016, 76, 75–96. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Diakumaku, E.; Krumbein, W.E.; (Geomicrobiology, Carl von Ossietzky University, Oldenburg, Germany). Personal communication, 1996.
- Selbmann, L.; Zucconi, L.; Isola, D.; Onofri, S. Rock black fungi: Excellence in the extremes. From the Antarctic to Space. Curr. Genet. 2015, 61, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; Onofri, S.; Zucconi, L.; Isola, D.; Rottigni, M.; Ghiglione, C.; Piazza, P.; Alvaro, M.C.; Schiaparelli, S. Distributional records of Antarctic fungi based on strains preserved in the Culture Collection of Fungi from Extreme Environments (CCFEE) Mycological Section associated with the Italian National Antarctic Museum (MNA). MycoKeys 2015, 10, 57–71. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Gebhardt, C.; Toepel, J.; Barry, J.; Munzke, N.; Günster, J.; Gorbushina, A.A. Monitoring microbial soiling in photovoltaic systems: A qPCR-based approach. Int. Biodeterior. Biodegrad. 2018, 129, 13–22. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Krumbein, W.E.; Hamman, C.H.; Panina, L.; Soukharjevski, S.; Wollenzien, U. Role of black fungi in color change and biodeterioration of antique marbles. Geomicrobiol. J. 1993, 11, 205–221. [Google Scholar] [CrossRef]
- Sterflinger, K.; De Baere, R.; De Hoog, G.S.; De Wachter, R.; Krumbein, W.E.; Haase, G. Coniosporium perforans and C. apollinis, two new rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie van Leeuwenhoek 1997, 72, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A.; Krumbein, W.E.; Volkmann, M. Rock surfaces as life indicators: New ways to demonstrate life and traces of former life. Astrobiology 2002, 2, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Dornieden, T.; Gorbushina, A.A.; Krumbein, W.E. Aenderungen der physikalischen Eigenschaften von Marmor durch Pilzbewuchs. Int. J. Restor. Build. Monum. 1997, 3, 441–456. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4239–4246. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA Polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6, Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Villesen 2017. Available online: http://users-birc.au.dk/biopv/php/fabox/alignment_joiner.php (accessed on 22 November 2017).
- Nylander, J.A.A. Mr AIC.pl. Programme Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Jobb, G.A.; von Haeseler, A.; Strimmer, K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 2004, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 40, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Plemenitaš, A.; Gunde-Cimerman, N. Cellular responses in the halophilic black yeast Hortaea werneckii to high environmental salinity. In Adaptation to Life at High Salt Concentrations in Archaea, Bacteria, and Eukarya; Gunde-Cimerman, N., Oren, A., Plemenitaš, A., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 455–470. [Google Scholar]
- Selbmann, L.; de Hoog, G.S.; Mazzaglia, A.; Friedmann, E.I.; Onofri, S. Fungi at the edge of life: Cryptoendolithic black fungi from Antarctic deserts. Stud. Mycol. 2005, 51, 1–32. [Google Scholar]
- Selbmann, L.; de Hoog, G.S.; Zucconi, L.; Isola, D.; Ruisi, S.; Gerrits van den Ende, A.H.G.; Ruibal, C.; De Leo, F.; Urzì, C.; Onofri, S. Drought meets acid: Three new genera in a dothidealean clade of extremotolerant fungi. Stud. Mycol. 2008, 61, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; Isola, D.; Egidi, E.; Zucconi, L.; Gueidan, C.; de Hoog, G.S.; Onofri, S. Mountain tips as reservoirs for new rock-fungal entities: Saxomyces gen. nov. and four new species from the Alps. Fungal Divers. 2014, 65, 167–182. [Google Scholar] [CrossRef]
- Egidi, E.; de Hoog, G.S.; Isola, D.; Onofri, S.; Quaedvlieg, W.; de Vries, M.; Verkley, G.J.M.; Stielow, J.B.; Zucconi, L.; Selbmann, L. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the dothidemycetes based on multi-locus phylogenies. Fungal Divers. 2014, 65, 127–165. [Google Scholar] [CrossRef]
- Sterflinger, K. Black Yeasts and Meristematic Fungi: Ecology, Diversity and Identification. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 501–514. [Google Scholar]
- Selbmann, L.; Isola, D.; Zucconi, L.; Onofri, S. Resistance to UV-B induced DNA damage in extreme-tolerant cryptoendolithic Antarctic fungi: Detection by PCR assays. Fungal Biol. 2011, 115, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, K.; Tesei, D.; Marzban, G.; Dijksterhuis, J.; Wyatt, T.; Sterflinger, K. Microcolonial fungi on rocks: A life in constant drought? Mycopathologia 2013, 175, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Selbmann, L.; Zucconi, L.; Raguse, M.; Moeller, R.; Shuryak, I.; Onofri, S. Survival, DNA integrity and ultrastructural damage in Antarctic cryptoendolithic eukaryotic microorganisms exposed to ionizing radiation. Astrobiology 2017, 17, 126–135. [Google Scholar] [CrossRef] [PubMed]


| Species | Strain | Origin | Location | GenBank Accession Number | ||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | ||||
| Austroafricana parva | CBS122892 = CPC 12421 EET | Eucalyptus globulus | Australia | KF901514 | KF901832 | KF902193 |
| Austroafricana parva | CBS110503 = CMW 14459 | Eucalyptus globulus | Australia | KF901513 | KF901831 | KF902189 |
| Austroafricana parva | CPC2120 | Protea repens | South Africa | AY260091 | - | - |
| Batcheloromyces proteae | CBS110696 = CPC1518 = CPC18701 | Protea cynaroides | South Africa | - | KF901833 | KF902195 |
| Constantinomyces macerans | CBS119304 = TRN440 T | Granite, Patones, Central Mountain System | Patones, Spain | AY843139 | KF310005 | KF310081 |
| Constantinomyces nebulosus | CBS117941 = TRN262 T | Granite, Atazar, Central Mountain System | Atazar, Spain | AY843109 | KF310014 | KF310068 |
| Constantinomyces oldenburgensisT | CBS144642 = CCFEE6311 = T2.1 | Photocatalytically active roof tiles | Edewecht, Germany | LT976552 * | LT976552 * | LT976526 |
| Constantinomyces oldenburgensis | CBS144643 = CCFEE6310 = T2.3 | Photocatalytically active roof tiles | Edewecht, Germany | LT976553 * | LT976553 * | LT976527 |
| Constantinomyces oldenburgensis | CBS144644 = CCFEE6309 = T2.4 | Photocatalytically active roof tiles | Edewecht, Germany | LT976554 * | LT976554 * | LT976528 |
| Constantinomyces oldenburgensis | CBS144645 = CCFEE6305 = T2.5 | Photocatalytically active roof tiles | Edewecht, Germany | LT976555 * | LT976555 * | LT976529 |
| Constantinomyces minimus | CBS118766 = TRN159 | Granite, La Cabrera, Central Mountain System | La Cabrera, Spain | AY843066 | KF310003 | KF310077 |
| Constantinomyces patonensisT | CBS117950 = TRN431 | Granite, Patones, Central Mountain System | Patones, Spain | AY843129 | KF310004 | KF310080 |
| Constantinomyces virgultus | CBS117930 = TRN79 T | Limestone, Cala Sant Vicenç | Mallorca, Spain | AY559339 | GU323964 | KF310082 |
| Elasticomyces elasticus | CBS122539 = CCFEE5319 | Lecanora sp. | Antarctica | FJ415475 | GU250375 | Unpublished |
| Elasticomyces elasticus | CBS122540 = CCFEE5320 | Usnea antarctica | Antarctica | FJ415476 | GU250376 | - |
| Friedmanniomyces endolithicus | CCFEE5199 | Rock | Antarctica | JN885547 | KF310007 | KF310093 |
| Friedmanniomyces endolithicus | CCFEE5180 | Rock | Antarctica | JN885544 | GU250367 | - |
| Friedmanniomyces endolithicus | CBS119426 = CCFEE670 | Rock | Antarctica | JN885542 | GU250366 | KF310056 |
| Friedmanniomyces endolithicus | CBS119427 = CCFEE 524 | Rock | Antarctica | JN885541 | GU250364 | KF310054 |
| Hortaea werneckii | CBS107.67 T | Tinea nigra: man | Portugal | AJ238468 | EU019270 | - |
| Hortaea werneckii | CBS117.90 | Salted fish, Osteoglossum bicirrhosum | Brazil | AJ238472 | - | - |
| Hortaea werneckii | CBS117931 = TRN122 | Rock | Cala Sant Vicenç, Mallorca, Spain | AY559357 | GU323969 | KF310058 |
| Hortaea werneckii | CBS110352 | Hollow tree | Karthoum | Unpublished | - | - |
| Incertomyces perditus | CBS136105 = CCFEE 5385 T | Rock | Alps, Italy | KF309977 | KF310008 | KF309977 |
| Incertomyces vagans | CCFEE5393 T | Rock | Alps, Italy | KF309964 | KF310009 | KF309964 |
| Lapidomyces hispanicus | CBS118355 = TRN500 | Rock | Puebla la Sierra, Spain | AY843182 | KF310017 | - |
| Lapidomyces hispanicus | CBS118764 = TRN126 | Cala Sant Vicenç | Mallorca, Spain | AY559361 | KF310016 | KF310076 |
| Meristemomyces frigidus | CBS136044 = CCFEE5401 T | Rock | Alps, Italy | KF309966 | GU250383 | KF310105 |
| Monticola elongata | CCFEE5499 | Rock | Alps, Italy | KF309969 | GU250398 | KF310065 |
| Monticola elongata | CBS137180 = CCFEE 5492 | Rock | Alps, Italy | KF309968 | KF309994 | KF310064 |
| Monticola elongata | CBS136206 = CCFEE5394 T | Rock | Alps, Italy | KF309965 | KF309995 | KF309965 |
| Neocatenulostroma abietis | CBS145.97 | Sandstone of cathedral | Germany | AJ244265 | - | - |
| Neocatenulostroma abietis | CBS290.90 | Skin lesion | Netherlands | AJ244267 | - | - |
| Neocatenulostroma microsporum | CBS110890 = CPC1832 ET | Protea cynaroides | South Africa | AY260097 | EU019255 | - |
| Neocatenulostroma microsporum | CBS101951 = CPC1960 ET | Protea cynaroides | South Africa | AY260097 | EU019255 | - |
| Oleoguttula mirabilis | CBS136101 = CCFEE5522 | Rock | Antarctic Peninsula | KF309972 | KF310019 | KF310070 |
| Oleoguttula mirabilis | CBS136102 = CCFEE5523 T | Rock | Antarctic Peninsula | KF309973 | KF310031 | Unpublished |
| Pseudoteratosphaeria ohnowa | CBS112896 = CPC1004 ET | Eucalyptus grandis | South Africa | KF901620 | KF901946 | KF902348 |
| Pseudoteratosphaeria ohnowa | CPC1005 | Eucalyptus grandis | South Africa | AF173299 | GU214511 | - |
| Pseudoteratosphaeria ohnowa | CBS113290 = CMW9102 | Eucalyptus smithii | South Africa | KF937236 | - | KF937270 |
| Pseudoteratosphaeria ohnowa | CBS110949 = CPC1006 | Eucalyptus grandis | South Africa | AY725575 | - | - |
| Pseudotaeniolina globosa | CBS109889 T | Rock | Italy | NR136960 | EU019283 | - |
| Pseudotaeniolina globosa | CBS303.84 | Rock | Italy | AJ244268 | - | - |
| Pseudotaeniolina globosa | CBS110353 | Human aorta at autopsy | Germany | Unpublished | - | - |
| Recurvomyces mirabilis | CBS119434 = CCFEE5264 T | Sandstone | Antarctica | FJ415477 | GU250372 | KF310059 |
| Recurvomyces sp. | CBS117957 = TRN491 | Quarzite | Puebla de la Sierra, Spain | AY1843175 | - | - |
| Suberoteratosphaeria suberosa | CBS11891 = CPC12085 ET | Eucalyptus sp. | Uruguay | KF901786 | KF902144 | - |
| Suberoteratosphaeria suberosa | CBS436.92 = CPC515 ET | Eucalyptus dunnii | Brazil | KF901623 | KF901949 | KF902404 |
| Teratosphaeria destructans | CBS111370 = CPC1368 ET | Eucalyptus grandis | Indonesia | KF901574 | KF901898 | KF902427 |
| Teratosphaeria eucalypti | CBS111692 = CMW14910 = CPC1582 | Eucalyptus sp. | New Zealand | - | KF902119 | - |
| Teratosphaeria eucalypti | CPC 12552 | Eucalyptus nitens | Tasmania | KF901576 | KF901900 | KF902429 |
| Teratosphaeria fibrillosa | CBS121707 = CPC13960 EET | Protea sp. | South Africa | KF901728 | KF902075 | - |
| Teratosphaeria macowanii | CBS122901 = CPC13899 EET | Protea nitida | South Africa | KF937241 | - | KF937276 |
| Teratosphaeria maxii | CBS120137 = CPC12805 ET | Protea repens | South Africa | KF937243 | - | KF937278 |
| Teratosphaeria maxii | CBS112496 = CPC3322 | Protea sp. | Australia | - | KF937242 | KF937277 |
| Teratosphaeria molleriana | CBS118359 = CMW11560 | Eucalyptus globulus | Tasmania | KF901764 | KF902120 | KF902451 |
| Teratosphaeria tinara | CBS124583 = MUCC666 ET | Corymbia sp. | Australia | KF901599 | KF901923 | KF902491 |
| Teratosphaeria toledana | CBS113313 = CMW14457 ET | Eucalyptus sp | Spain | KF901734 | KF902081 | KF902492 |
| Unidentified rock inhabiting fungus | CCFEE507 | Powdered rocks | Antarctica | Unpublished | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruibal, C.; Selbmann, L.; Avci, S.; Martin-Sanchez, P.M.; Gorbushina, A.A. Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces. Life 2018, 8, 30. https://doi.org/10.3390/life8030030
Ruibal C, Selbmann L, Avci S, Martin-Sanchez PM, Gorbushina AA. Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces. Life. 2018; 8(3):30. https://doi.org/10.3390/life8030030
Chicago/Turabian StyleRuibal, Constantino, Laura Selbmann, Serap Avci, Pedro M. Martin-Sanchez, and Anna A. Gorbushina. 2018. "Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces" Life 8, no. 3: 30. https://doi.org/10.3390/life8030030
APA StyleRuibal, C., Selbmann, L., Avci, S., Martin-Sanchez, P. M., & Gorbushina, A. A. (2018). Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces. Life, 8(3), 30. https://doi.org/10.3390/life8030030







