CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity
Abstract
:1. Introduction
2. Viruses and Conjugative Plasmids of the Sulfolobales
2.1. Viruses
| Family | Name | Host | Morphotype | Genome | Integrates | Genome Size |
|---|---|---|---|---|---|---|
| Fuselloviridae | ASV1 | Acidianus | Fusiform | Circular | yes | 24,655 |
| SSV1, SSV2, | Sulfolobus | 15,465, 14,796 | ||||
| SSV4, SSV5, | " | 15,135, 15,330 | ||||
| SSV6, SSV7 | " | 15,684, 17,602 | ||||
| SMF1 | Sulfolobales | 14,847 | ||||
| Icosahedral | STIV | Sulfolobus | Turreted icosahedral | Circular | no | 17,663 |
| STIV2 | " | 16,622 | ||||
| Bicaudaviridae | ATV, ATV2 | Acidianus | Tailed-fusiform | Circular | yes | 62,730, 57,909 |
| SMV1 | Sulfolobus | 48,775 | ||||
| Monocauda-viruses | STSV1 | Sulfolobus | Tailed-fusiform | Circular | yes | 75,294 |
| STSV2 | " | 76,107 | ||||
| Rudiviridae | ARV1 | Acidianus | Rod-shaped | Linear | no | 24,655 |
| SIRV1, SIRV2 | Sulfolobus | 32,308, 35,450 | ||||
| SIRV4 | " | 32,992 | ||||
| SMR1 | Sulfolobales | 27,431 | ||||
| Lipothrixviridae | AFV1, AFV2 | Acidianus | Filamentous | Linear | no | 20,869, 31,787 |
| AFV3, AFV6 | " | 40,449, 39,577 | ||||
| AFV7, AFV8 | " | 36,895, 38,179 | ||||
| AFV9 | " | 41,172 | ||||
| SIFV, SIFV2 | Sulfolobus | 40,900, 39,399 | ||||
| Ampullaviridae | ABV | Acidianus | Bottle-shaped | Linear | no | 23,814 |
| Guttaviridae | SNDV | Sulfolobus | Bearded droplet | Circular(modified) | - | unsequenced |
2.2. Conjugative Plasmids
| Plasmid | Host | Origin | Genome size |
|---|---|---|---|
| pARN3 | S. islandicus | Iceland | 26,200 |
| pARN4 | " | " | 26,476 |
| pHVE14 | " | " | 35,422 |
| pING1 | " | " | 24,554 |
| pKEF9 | " | " | 28,930 |
| pSOG1 | " | " | 29,000 |
| pSOG2 | " | " | 25,960 |
| pAH1 | Acidianus hospitalis | Italy | 28,649 |
| pMGB1 | S. solfataricus | USA | 27,975 |
| pNOB8 | Sulfolobus sp. NOB8H2 | Japan | 41,229 |
| pTC | S. tengchongensis | China | 20,417 |
3. Different Classes of CRISPR-Cas Systems
3.1. Structural Classification
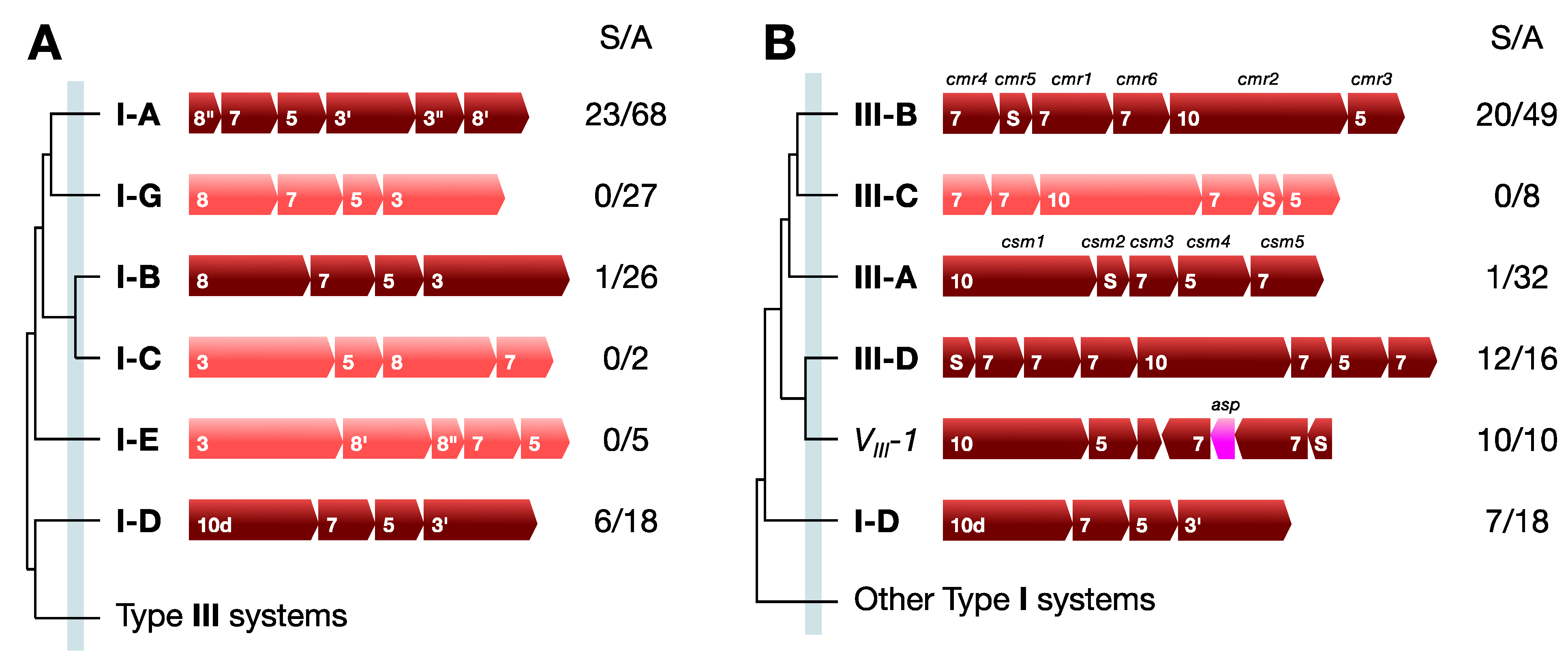
3.2. Functional Classification
4. Properties of CRISPR Loci
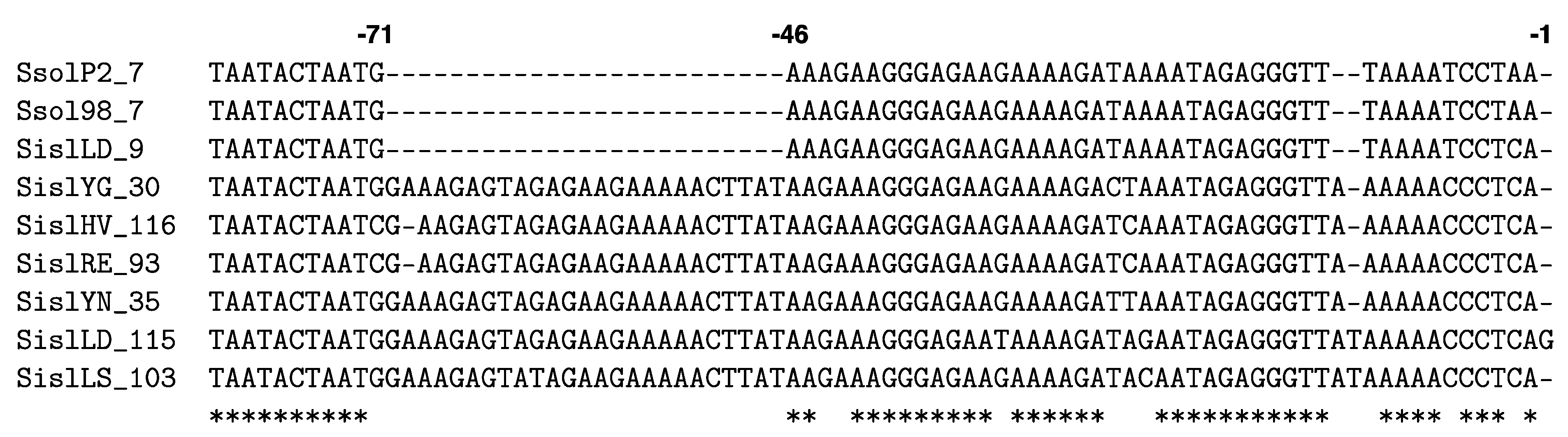
4.1. Repeats, Spacers and Leaders

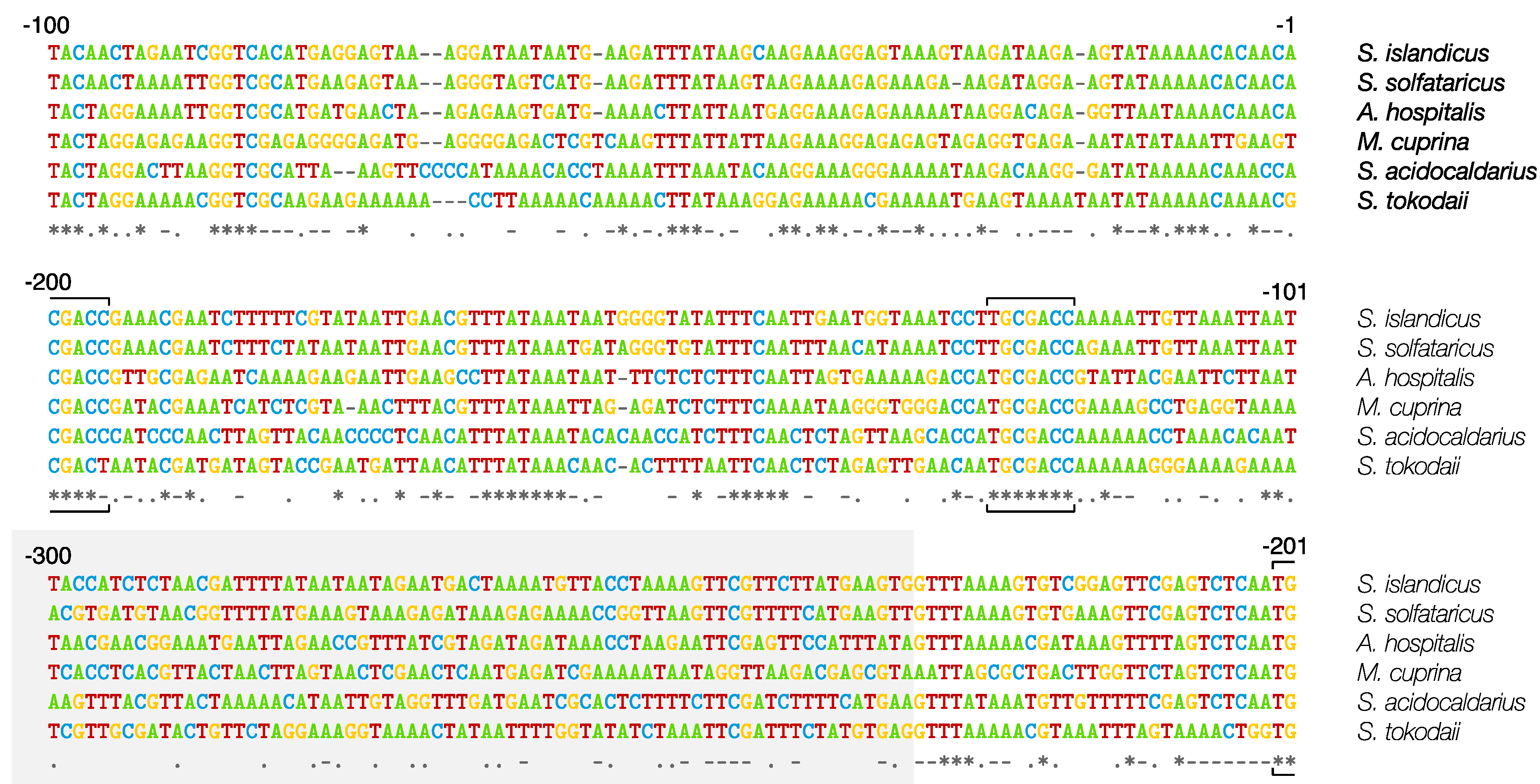
4.2. Spacer Sequence Matches to Invasive Genetic Elements
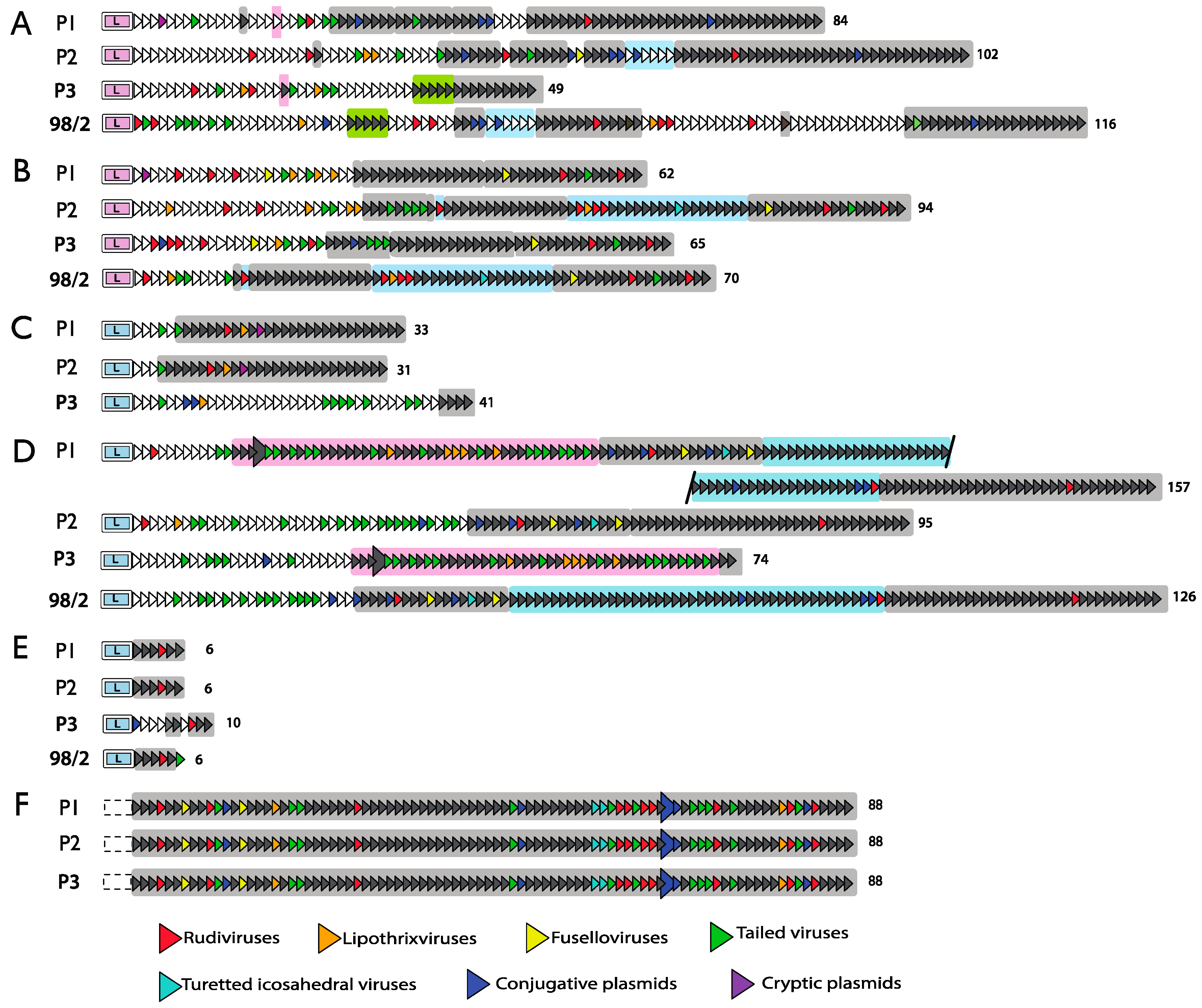
4.3. Transcription and Processing
4.4. Structural Instability of CRISPR Loci
4.5. Integrity of the Spacer-Repeat Substructure
5. Modular Mechanisms of Adaptation
5.1. Adaptation Module
5.2. Mechanism of Protospacer Selection
5.3. Exceptional Biased Spacer Selection from a Conjugative Plasmid
5.4. De Novo Spacer Insertion
5.5. Alternative Spacer Acquisition Mechanism
5.6. Is Adaptation Activated by Interference?
5.7. The Conundrum of Reversible De Novo Spacer Acquisition
6. Molecular Mechanisms of Interference
6.1. Functional Significance of the Strand-Specificity of Spacer Matches
6.2. Fidelity of crRNA-Spacer Recognition
6.3. Unequal Assembly of crRNAs into Interference Complexes
6.4. Type I PAM-Dependent Interference
6.5. Type III PAM-Independent Interference
6.6. Quaternary Structures of Interference Complexes
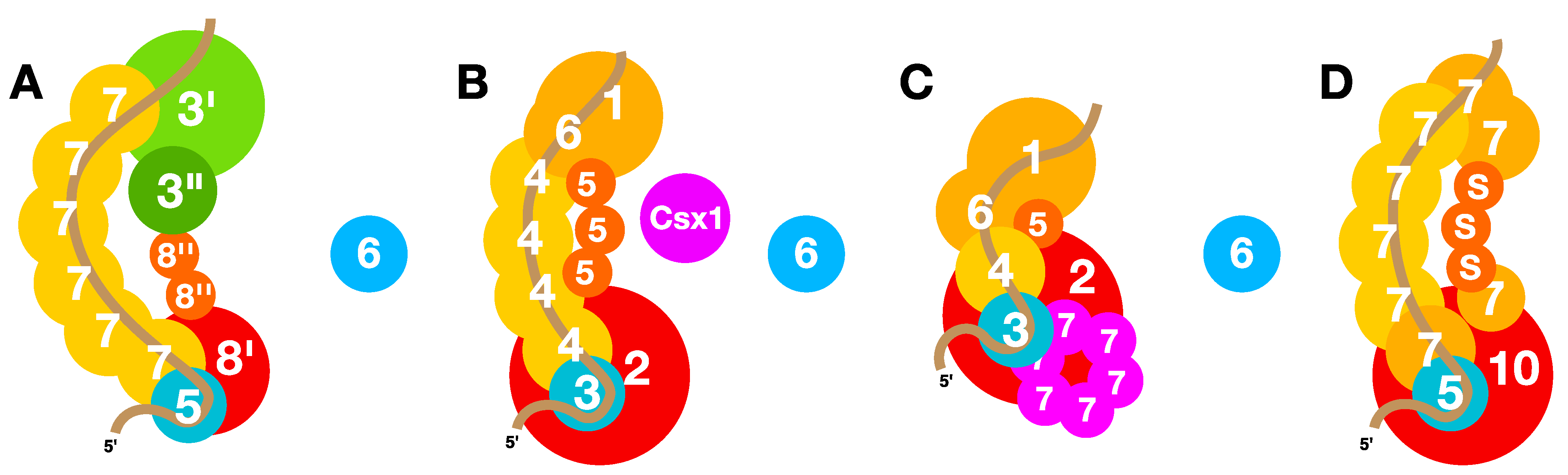
7. Inhibitory and Regulatory Mechanisms of Adaptation and Interference
7.1. Differential Regulation of Adaptation and Interference
7.2. Inactivation of a Regulatory cas Gene by Genetic Element Integration
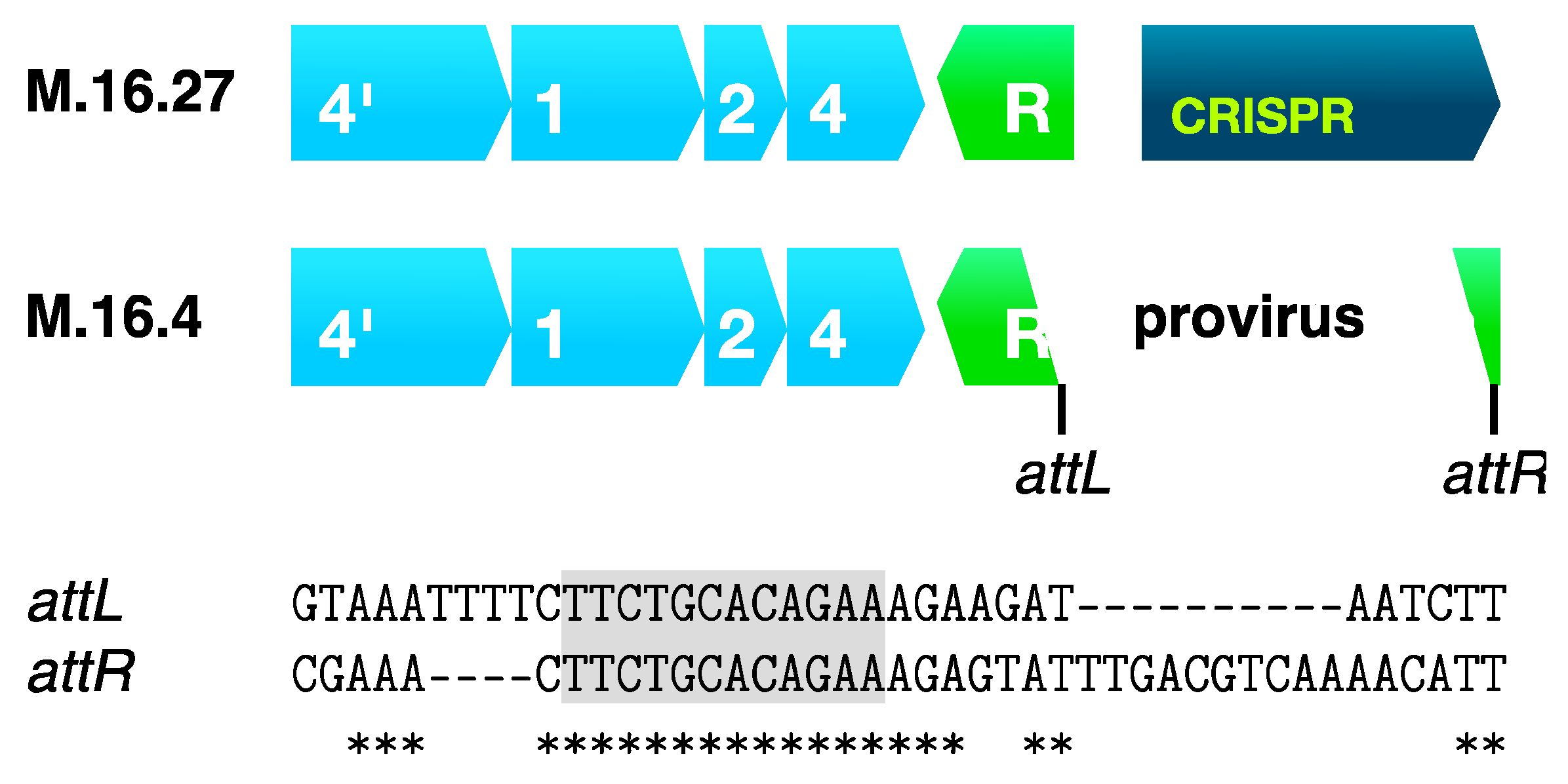
7.3. CRISPR-Cas Interference Avoidance and Anti-CRISPR Systems
7.4. Integration and Interference
7.5. CRISPR-Cas Defence and Transposition
7.6. Antisense CRISPR RNAs Can Impair Interference Effects
8. Role for Toxins
9. Functional Importance of Non-Core Cas Proteins
9.1. Csx1 Superfamily Proteins
9.2. Proteases
9.3. ATPases
9.4. CRISPR Repeat Binding Proteins Cbp1 and Cbp2
10. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- She, Q.; Singh, R.K.; Confalonieri, F.; Zivanovic, Y.; Gordon, P.; Allard, G.; Awayez, M.J.; Chan-Weiher, C.-Y.; Clausen, I.G.; Curtis, B.; et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 2001, 98, 7835–7840. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Arnold, H.P.; Holz, I.; Prangishvili, D.; Schweier, A.; Stedman, K.; She, Q.; Phan, H.; Garrett, R.A.; Kristjansson, J.K. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 1998, 2, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.M.; Garrett, R.A. Discovery and seminal developments in the CRISPR field. In CRISPR-Cas Systems; Barrangou, R., van der Oost, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–31. [Google Scholar]
- Manica, A.; Schleper, C. CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA Biol. 2013, 10, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Jonuscheit, M.; Martusewitsch, E.; Stedman, K.M.; Schleper, C. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 2003, 48, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Zhang, C.; Deng, L.; Peng, N.; Chen, Z.; Liang, Y.X. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 2009, 37, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhu, H.; Chen, Z.; Liang, Y.X.; She, Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 2009, 13, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, L.; Deng, L.; Wu, Y.; Liang, Y.; Huang, L.; She, Q. Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology 2010, 156, 3386–3397. [Google Scholar] [CrossRef] [PubMed]
- Manica, A.; Zebec, Z.; Teichmann, D.; Schleper, C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 2011, 80, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Peng, N.; Deng, L.; Mei, Y.; Jiang, D.; Hu, Y.; Awayez, M.; Liang, Y.; She, Q. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012, 78, 5630–5637. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; van Wolferen, M.; Wagner, A.; Lassak, K.; Meyer, B.H.; Reimann, J.; Albers, S.V. Versatile Genetic Tool Box for the Crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Zhang, C.; Krause, D.J.; Whitaker, R.J. Sulfolobus islandicus: A model system for evolutionary genomics. Biochem. Soc. Trans. 2013, 41, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Forterre, P.; Garrett, R.A. Viruses of the Archaea: A unifying view. Nat. Rev. Microbiol. 2006, 4, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Pina, M.; Bize, A.; Forterre, P.; Prangishvili, D. The archaeoviruses. FEMS Microbiol. Rev. 2011, 35, 1035–1054. [Google Scholar] [CrossRef] [PubMed]
- Luk, A.W.S.; Williams, T.J.; Erdmann, S.; Papke, R.T.; Cavicchioli, R. Viruses of haloarchaea. Life 2014, 4, 681–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, N.; Shah, S.A.; Huang, L.; She, Q. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 2015, 79, 117–152. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, B.; Shaughnessy, D.P.; Wolf, Y.I.; Koonin, E.V.; Roberto, F.F.; Young, M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012, 86, 5562–5573. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Erdmann, S.; She, Q.; Garrett, R.A. Novel virus-host interactions in Sulfolobus. Manuscript under preparation.
- Schleper, C.; Holz, I.; Janekovic, D.; Murphy, J.; Zillig, W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 1995, 177, 4417–4426. [Google Scholar] [PubMed]
- She, Q.; Phan, H.; Garrett, R.A.; Albers, S-V.; Stedman, K.M.; Zillig, W. Genetic profile of pNOB8 from Sulfolobus: The first conjugative plasmid from an archaeon. Extremophiles 1998, 2, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Albers, S.-V.; Holz, I.; Arnold, H.P.; Stedman, K.; Klein, T.; Singh, H.; Hiort, J.; Schweier, A.; Kristjansson, J.K.; et al. Conjugation in archaea: Frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 1998, 40, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Stedman, K.M.; She, Q.; Phan, H.; Holz, I.; Singh, H.; Prangishvili, D.; Garrett, R.; Zillig, W. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: Insights into recombination and conjugation in Crenarchaeota. J. Bacteriol. 2000, 182, 7014–7020. [Google Scholar] [CrossRef] [PubMed]
- Greve, B.; Jensen, S.; Brügger, K.; Zillig, W.; Garrett, R.A. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea 2004, 1, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Erauso, G.; Stedman, K.M.; van de Werken, H.J.; Zillig, W.; van der Oost, J. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology 2006, 152, 1951–1968. [Google Scholar] [CrossRef] [PubMed]
- Basta, T.; Smyth, J.; Forterre, P.; Prangishvili, D.; Peng, X. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 2009, 71, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Lillestøl, R.K.; Redder, P.; Garrett, R.A.; Brügger, K. A putative viral defence mechanism in archaeal cells. Archaea 2006, 2, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Brügger, K.; Skovgaard, M.; Redder, P.; She, Q.; Torarinsson, E.; Greve, B.; Awayez, M.; Zibat, A.; Klenk, H.-P.; et al. The Genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005, 187, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Garrett, R.A. RNA-protein interactions of an archaeal homotetrameric splicing endonuclease with an exceptional evolutionary history. EMBO J. 1997, 16, 6290–6300. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Rozhdestvensky, T.S.; d’Orval, B.C.; Bortolin, M.L.; Huber, H.; Charpentier, B.; Branlant, C.; Bachellerie, J.P.; Brosius, J.; Hüttenhofer, A. RNomics in archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002, 30, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Koonin, E.V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 2011, 6. [Google Scholar] [CrossRef]
- Vestergaard, G.; Garrett, R.A.; Shah, S.A. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014, 11, 157–168. [Google Scholar] [CrossRef]
- Lillestøl, R.K.; Shah, S.A.; Brügger, K.; Redder, P.; Phan, H.; Christiansen, J.; Garrett, R.A. CRISPR families of the crenarchaeal genus Sulfolobus: Bidirectional transcription and dynamic properties. Mol. Microbiol. 2009, 72, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Garrett, R.A. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011, 162, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Garrett, R.A.; Vestergaard, G.; Shah, S.A. Archaeal CRISPR-based immune systems: Exchangeable functional modules. Trends Microbiol. 2011, 19, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Almendros, C.; Mojica, F.J.M.; Díez-Villaseñor, C.; Guzmán, N.M.; García-Martínez, J. CRISPR-Cas functional module exchange in Escherichia coli. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005, 1. [Google Scholar] [CrossRef]
- Garrett, R.A.; Shah, S.A.; Vestergaard, G.; Deng, L.; Gudbergsdottir, S.; Kenchappa, C.S.; Erdmann, S.; She, Q. CRISPR-based immune systems of the Sulfolobales: Complexity and diversity. Biochem. Soc. Trans. 2011, 39, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rouillon, C.; Kerou, M.; Reeks, J.; Brügger, K.; Graham, S.; Reimann, J.; Cannone, G.; Liu, H.; Albers, S.V.; Naismith, J.H.; Spagnolo, L. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 2012, 45, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zebec, Z.; Manica, A.; Zhang, J.; White, M.F.; Schleper, C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014, 42, 5280–5288. [Google Scholar] [CrossRef] [PubMed]
- Staals, R.H.; Agari, Y.; Maki-Yonekura, S.; Zhu, Y.; Taylor, D.W.; van Duijn, E.; Barendregt, A.; Vlot, M.; Koehorst, J.J.; Sakamoto, K.; et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell 2013, 52, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Garrett, R.A.; Shah, S.A.; Peng, X.; She, Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013, 87, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Feng, M.; Feng, X.; Liang, Y.X.; She, Q. An archaeal CRISPR type III-B Cmr system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015, 43, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, G.W.; Jiang, W.; Bikard, D.; Marraffini, L.A. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 2014, 514, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Live virus-free or die: Coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 2012, 7. [Google Scholar] [CrossRef]
- Lange, S.J.; Alkhnbashi, O.S.; Rose, D.; Will, S.; Backofen, R. CRISPRmap: An automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 2013, 41, 8034–8044. [Google Scholar] [CrossRef] [PubMed]
- Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Garrett, R.A.; Saunders, S.J.; Backofen, R. CRISPRstrand: Predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 2014, 30, i489–i496. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Hansen, N.R.; Garrett, R.A. Distributions of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Trans. Biochem. Soc. 2009, 37, 23–28. [Google Scholar] [CrossRef]
- Held, N.L.; Whitaker, R.J. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 2009, 11, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Brügger, K.; Liu, C.; Shah, S.A.; Zheng, H.; Zhu, Y.; Wang, S.; Lillestøl, R.K.; Chen, L.; Frank, J.; et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus- host interaction studies. J. Bacteriol. 2011, 193, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Vestergaard, G.; Garrett, R.A. CRISPR/Cas and CRISPR/Cmr Immune Systems of Archaea. In Regulatory RNAs in Prokaryotes; Marchfelder, A., Hess, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 163–181. [Google Scholar]
- Lillestøl, R.K. A Study of CRISPR-Systems and ncRNAs in Sulfolobus. PhD Thesis, University of Copenhagen, Copenhagen, Denmark, 2009. [Google Scholar]
- Jaubert, C.; Danioux, C.; Oberto, J.; Cortez, D.; Bize, A.; Krupovic, M.; She, Q.; Forterre, P.; Prangishvili, D.; Sezonov, G. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 2013, 3. [Google Scholar] [CrossRef]
- Shah, S.A.; Erdmann, S.; Mojica, F.J.; Garrett, R.A. Protospacer motifs: Mixed identities and functional diversity. RNA Biol. 2013, 10, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Bachellerie, J.P.; Rozhdestvensky, T.; Bortolin, M.L.; Huber, H.; Drungowski, M.; Elge, T.; Brosius, J.; Hüttenhofer, A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 2002, 99, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Polacek, N.; Zywicki, M.; Huber, H.; Brügger, K.; Garrett, R.A.; Bachellerie, J.P.; Hüttenhofer, A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; Kerou, M.; Brumfield, S.K.; Graham, S.; Liu, H.; Naismith, J.H.; Sdano, M.; Peng, N.; She, Q.; Copie, V.; et al. Structural and functional characterization of an archaeal CASCADE complex for CRISPR-mediated viral defense. J. Biol. Chem. 2011, 286, 21643–21656. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Li, H.; Hallstrøm, S.; Peng, N.; Liang, Y.X.; She, Q. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013, 10, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Reeks, J.; Sokolowski, R.D.; Graham, S.; Liu, H.; Naismith, J.H.; White, M.F. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem. J. 2013, 452, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, R.D.; Graham, S.; White, M.F. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res. 2014, 42, 6532–6541. [Google Scholar] [CrossRef] [PubMed]
- Brendel, J.; Stoll, B.; Lange, S.J.; Sharma, K.; Lenz, C.; Stachler, A.E.; Maier, L.K.; Rihter, H.; Schmitz, R.A.; Randau, L.; et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J. Biol. Chem. 2014, 289, 7164–7177. [Google Scholar] [CrossRef] [PubMed]
- Torarinsson, E.; Klenk, H.-P.; Garrett, R.A. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 2005, 7, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.W.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Kenchappa, C.S.; Peng, X.; She, Q.; Garrett, R.A. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012, 40, 2470–2480. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Chen, B.; Huang, X.; Deng, L.; Liu, C.; Shah, S.A.; Le Moine Bauer, S.; Sobrino, C-L.; Wang, H.; Wei, Y.; et al. A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 2014, 18, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Le Moine Bauer, S.; Garrett, R.A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 2014, 91, 900–917. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W. Homologous recombination in Sulfolobus acidocaldarius: Genetic assays and functional properties. Biochem. Soc. Trans. 2009, 37, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Held, N.L.; Herrera, A.; Cadillo-Quiroz, H.; Whitaker, R.J. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Garrett, R.A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012, 85, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Shah, S.A.; Garrett, R.A. SMV1 virus-induced CRISPR spacer acquisition from the conjugative plasmid pMGB1 in Sulfolobus solfataricus P2. Biochem. Trans. Soc. 2013, 41, 1449–1458. [Google Scholar]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010, 26, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Gudbergsdottir, S.; Deng, L.; Chen, Z.; Jensen, J.V.K.; Jensen, L.R.; She, Q.; Garrett, R.A. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011, 79, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Brügger, K.; Redder, P.; She, Q.; Confalonieri, F.; Zivanovic, Y.; Garrett, R.A. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 2002, 206, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Brügger, K.; Torarinsson, E.; Chen, L.; Garrett, R.A. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 2004, 32, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Redder, P.; She, Q.; Garrett, R.A. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 2001, 306, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Redder, P.; Garrett, R.A. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 2006, 188, 4198–4206. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Beloglazova, N.; Flick, R.; Graham, C.; Skarina, T.; Nocek, B.; Gagarinova, A.; Pogoutse, O.; Brown, G.; et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011, 79, 484–502. [Google Scholar] [CrossRef] [PubMed]
- Paez-Espino, D.; Morovic, W.; Sun, C.L.; Thomas, B.C.; Ueda, K.; Stahl, B.; Barrangou, R.; Banfield, J.F. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576. [Google Scholar] [CrossRef] [PubMed]
- Díez-Villaseñor, C.; Guzmán, N.M.; Almendros, C.; García-Martínez, J.; Mojica, F.J.M. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 2013, 10, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Reno, M.L.; Held, N.L.; Fields, C.J.; Burke, P.V.; Whitaker, R.J. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. USA 2009, 106, 8605–8610. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Pougach, K.; Tikhonov, A.; Wanner, B.L.; Severinov, K.; Semenova, E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef]
- Swarts, D.C.; Mosterd, C.; van Passel, M.W.; Brouns, S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, R.; Zhao, D.; Xiang, H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014, 42, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.F.; Voet, M.; Sismeiro, O.; Dillies, M.-A.; Jagla, B.; Coppée, J.-Y.; Sezonov, G.; Forterre, P.; van der Oost, J.; Lavigne, R.; et al. Massive activation of archaeal defense genes during viral infection. J. Virol. 2013, 87, 8419–8428. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V.C., III; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Barrangou, R.; Garneau, J.E.; Labonté, J.; Fremaux, C.; Boyaval, P.; Romero, D.A.; Horvath, P.; Moineau, S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Romero, D.A.; Coûté-Monvoisin, A.C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Manica, A.; Zebec, Z.; Steinkellner, J.; Schleper, C. Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2013, 41, 10509–10517. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Mousaei, M.; Deng, L.; She, Q.; Garrett, R.A. crRNA-protospacer recognition during CRISPR-directed DNA interference in Sulfolobus. Manuscript under preparation.
- Rouillon, C.; Zhou, M.; Zhang, J.; Politis, A.; Beilsten-Edmands, V.; Cannone, G.; Graham, S.; Robinson, C.V.; Spagnolo, L.; White, M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 2013, 52, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Holz, I.; Zillig, W.; Garrett, R.A.; She, Q. Evolution of the family of pRN plasmids and their integrase-mediated insertion into the chromosome of the Crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 2000, 303, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Maier, L.K.; Stoll, B.; Brendel, J.; Fischer, E.; Pfeiffer, F.; Dyall-Smith, M.; Marchfelder, A. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012, 287, 33351–33363. [Google Scholar] [CrossRef] [PubMed]
- Plagens, A.; Tripp, V.; Daume, M.; Sharma, K.; Klingl, A.; Hrle, A.; Conti, E.; Urlaub, H.; Randau, L. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014, 42, 5125–5138. [Google Scholar] [CrossRef] [PubMed]
- Tamulaitis, G.; Kazlauskiene, M.; Manakova, E.; Venclovas, C.; Nwokeoji, A.O.; Dickman, M.J.; Horvath, P.; Siksnys, V. Programmable RNA shredding by the Type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 2014, 56, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. CRISPR-Cas Evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013, 10, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Lander, G.C.; Zhou, K.; Jore, M.M.; Brouns, S.J.; van der Oost, J.; Doudna, J.A.; Nogales, E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011, 477, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Spilman, M.; Cocozaki, A.; Hale, C.; Shao, Y.; Ramia, N.; Terns, R.; Terns, M.; Li, H.; Stagg, S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell 2013, 52, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Hatoum-Aslan, A.; Samai, P.; Maniv, I.; Jiang, W.; Marraffini, L.A. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013, 288, 27888–27897. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; Frankel, K.A.; Tsutakawa, S.E.; Alsbury, D.L.; Copié, V.; Young, M.J.; Tainer, J.A.; Lawrence, C.M. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J. Mol. Biol. 2011, 405, 939–955. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Wang, X.; Ye, Q.; Li, H.; Liang, Y.; She, Q.; Peng, N. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015, 43, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- León-Sobrino, C.; Kot, W.P.; Garrett, R.A. A transcriptome study of de novo spacer acquisition from the STSV2 virus by CRISPR loci of Sulfolobus islandicus REY15A. Manuscript under preparation.
- Pul, U.; Wurm, R.; Arslan, Z.; Geissen, R.; Hofmann, N.; Wagner, R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010, 75, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Pul, U.; Heidrich, N.; Jore, M.M.; Lundgren, M.; Stratmann, T.; Wurm, R.; Raine, A.; Mescher, M.; Mastop, M.; et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010, 77, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Scholz, I.; Voss, B.; Hess, W.R. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013, 10, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Kessler, A.; Phan, H.; Garrett, R.A.; Prangishvili, D. Multiple variants of the archaeal DNA rudivirus SIRV1 in a single host and a novel mechanism of genome variation. Mol. Microbiol. 2004, 54, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, G.; Shah, S.A.; Bize, A.; Reitberger, W.; Reuter, M.; Phan, H.; Briegel, A.; Rachel, R.; Garrett, R.A.; Prangishvili, D. SRV, a new rudiviral isolate from Stygioglobus and the interplay of crenarchaeal rudiviruses with the host viral-defence CRISPR system. J. Bacteriol. 2008, 190, 6837–6845. [Google Scholar] [CrossRef] [PubMed]
- Garrett, R.A.; Prangishvili, D.; Shah, S.A.; Reuter, M.; Stetter, K.; Peng, X. Metagenomic analyses of novel viruses, plasmids, and their variants, from an environmental sample of hyperthermophilic neutrophiles cultured in a bioreactor. Environ. Microbiol. 2010, 12, 2918–2930. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, G.; Aramayo, R.; Basta, T.; Häring, M.; Peng, X.; Brügger, K.; Chen, L.; Rachel, R.; Boisset, N.; Garrett, R.A.; et al. Structure of the Acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J. Virol. 2008, 82, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Redder, P.; Peng, X.; Brügger, K.; Shah, S.A.; Roesch, F.; Greve, B.; She, Q.; Schleper, C.; Forterre, P.; Garrett, R.A.; et al. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ. Microbiol. 2009, 11, 2849–2862. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.L.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2012, 493, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Bondy-Denomy, J.; Cheung, V.H.; Maxwell, K.L.; Davidson, A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Muskhelishvili, G.; Palm, P.; Zillig, W. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol. Gen. Genet. 1993, 237, 334–342. [Google Scholar] [PubMed]
- She, Q.; Shen, B.; Chen, L. Archaeal integrases and mechanisms of gene capture. Biochem. Soc. Trans. 2004, 32, 222–226. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Peng, X.; Zillig, W.; Garrett, R.A. Genome evolution: Gene capture events in archaeal chromosomes. Nature 2001, 409. [Google Scholar] [CrossRef] [PubMed]
- You, X.Y.; Liu, C.; Wang, S.Y.; Jiang, C.Y.; Shah, S.A.; Prangishvili, D.; She, Q.; Liu, S.J.; Garrett, R.A. Genomic studies of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles. Extremophiles 2011, 15, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Vestergaard, G.; Häring, M.; Aramayo, R.; Basta, T.; Rachel, R.; Garrett, R.A. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J. Mol. Biol. 2006, 359, 1203–1216. [Google Scholar] [CrossRef] [PubMed]
- Ortmann, A.C.; Brumfield, S.K.; Walther, J.; McInnerney, K.; Brouns, S.J.; van de Werken, H.J.; Bothner, B.; Douglas, T.; van der Oost, J.; Young, M.J. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 2008, 82, 4874–4883. [Google Scholar] [CrossRef] [PubMed]
- Okutan, E.; Deng, L.; Mirlashari, S.; Uldahl, K.; Halim, M.; Liu, C.; Garrett, R.A.; She, Q.; Peng, X. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol. 2013, 10, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Zago, M.A.; Dennis, P.P.; Omer, A.D. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 1812–1828. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1. [Google Scholar] [CrossRef]
- Shah, S.A.; Garrett, R.A. Archaeal type II toxin-antitoxins. In Prokaryotic Toxin-Antitoxins; Gerdes, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 225–238. [Google Scholar]
- Makarova, K.S.; Anantharaman, V.; Grishin, N.V.; Koonin, E.V.; Aravind, L. CARF and WYL domains: Ligand binding regulators of prokaryotic defense systems. Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- He, F.; Chen, L.; Peng, X. First experimental evidence for the presence of a CRISPR toxin in Sulfolobus. J. Mol. Biol. 2014, 426, 3683–3688. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Brügger, K.; Shen, B.; Chen, L.; She, Q.; Garrett, R.A. Genus-specific protein binding to the large clusters of DNA repeats (Short Regularly Spaced Repeats) present in Sulfolobus genomes. J. Bacteriol. 2003, 185, 2410–2417. [Google Scholar] [CrossRef] [PubMed]
- Kenchappa, C.S.; Heidarsson, P.O.; Kragelund, B.B.; Garrett, R.A.; Poulsen, F.M. Solution properties of the archaeal CRISPR DNA repeat-binding homeodomain protein Cbp2. Nucleic Acids Res. 2013, 41, 3424–3435. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrett, R.A.; Shah, S.A.; Erdmann, S.; Liu, G.; Mousaei, M.; León-Sobrino, C.; Peng, W.; Gudbergsdottir, S.; Deng, L.; Vestergaard, G.; et al. CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity. Life 2015, 5, 783-817. https://doi.org/10.3390/life5010783
Garrett RA, Shah SA, Erdmann S, Liu G, Mousaei M, León-Sobrino C, Peng W, Gudbergsdottir S, Deng L, Vestergaard G, et al. CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity. Life. 2015; 5(1):783-817. https://doi.org/10.3390/life5010783
Chicago/Turabian StyleGarrett, Roger A., Shiraz A. Shah, Susanne Erdmann, Guannan Liu, Marzieh Mousaei, Carlos León-Sobrino, Wenfang Peng, Soley Gudbergsdottir, Ling Deng, Gisle Vestergaard, and et al. 2015. "CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity" Life 5, no. 1: 783-817. https://doi.org/10.3390/life5010783
APA StyleGarrett, R. A., Shah, S. A., Erdmann, S., Liu, G., Mousaei, M., León-Sobrino, C., Peng, W., Gudbergsdottir, S., Deng, L., Vestergaard, G., Peng, X., & She, Q. (2015). CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity. Life, 5(1), 783-817. https://doi.org/10.3390/life5010783








