Prebiotic RNA Synthesis by Montmorillonite Catalysis
Abstract
:1. Introduction
2. Experimental Section
2.1. General
2.2. Analytical Methods
2.3. Preparation of Catalytic Montmorillonite
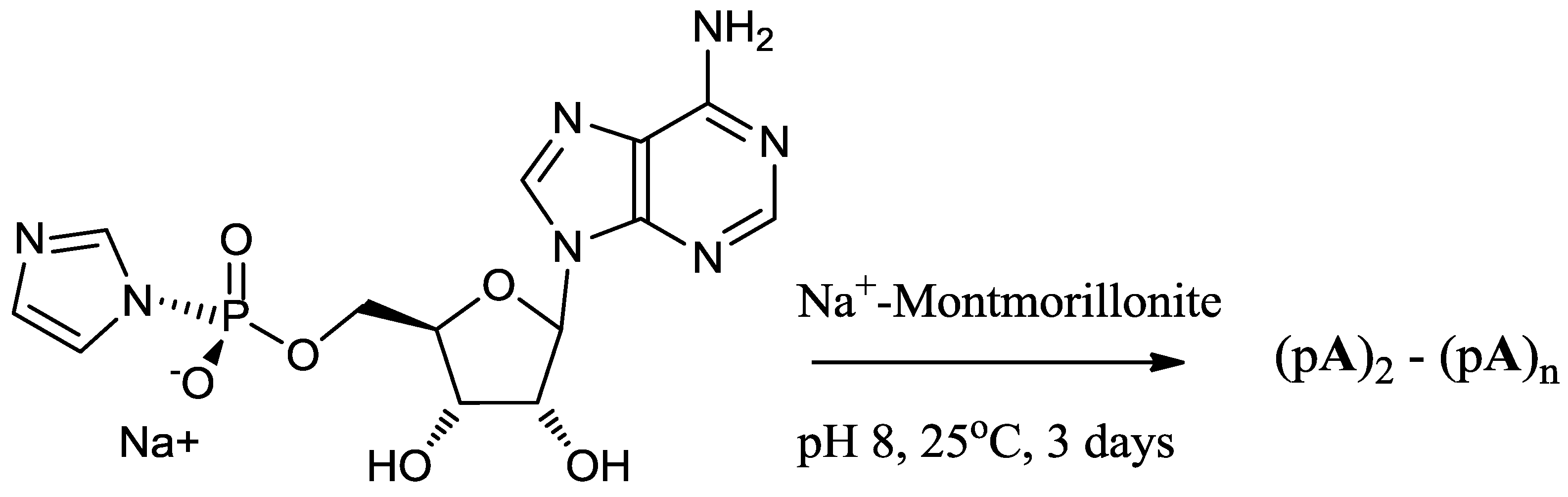
2.4. Preparation of the Activated Nucleotide of AMP

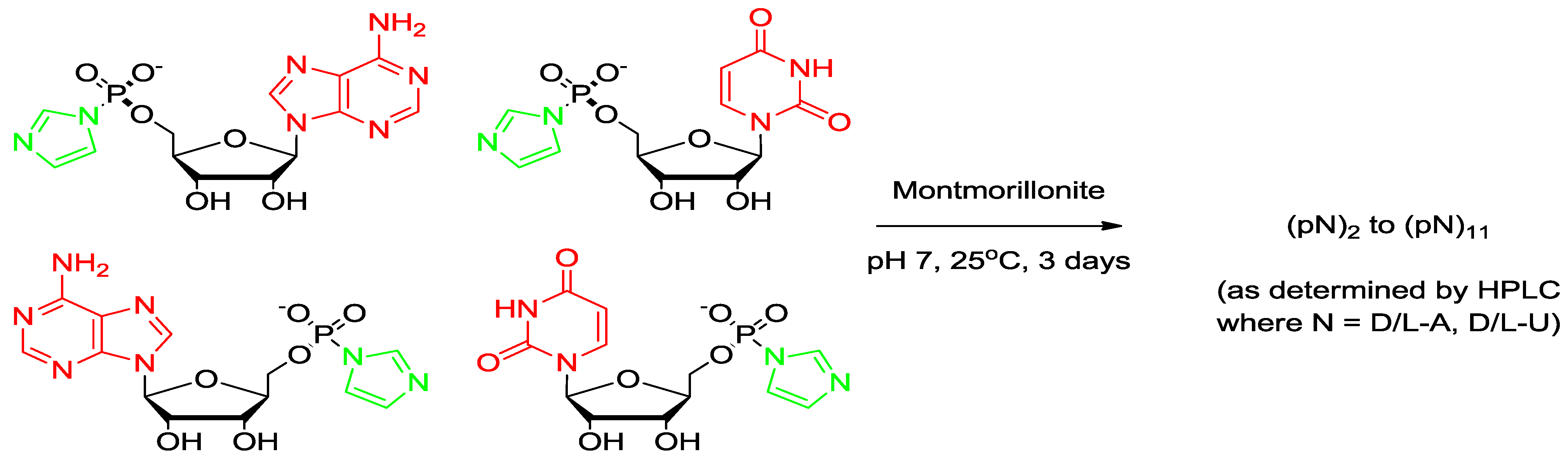
2.5. Montmorillonite-Catalyzed Oligomerization of ImpA
- Reaction 2. d-ImpA with d-ImpU (Total 15 mM) on Na+-montmorillonite; and
- Reaction 3. d,l-ImpA with d,l-ImpU (Total 15 mM) on Na+-montmorillonite.
2.6. Reaction and Analysis in the Absence of Minerals
2.7. Enzymatic Hydrolysis of Reaction Products
3. Results and Discussion
3.1. Significance of the Investigation
3.2. Oligomerization of Activated Mononucleotide on Na+-Montmorillonite
| Homochirality | Monomer | Dimer | Trimer | Tetramer | Pentamer |
|---|---|---|---|---|---|
| Observed | 50% | 63.5% | 74.3% | 92.7% | 97.2% |
| Calculated | 50% | 50% | 25% | 12.5% | 6.25% |
3.3. Analysis of the Products of Reaction
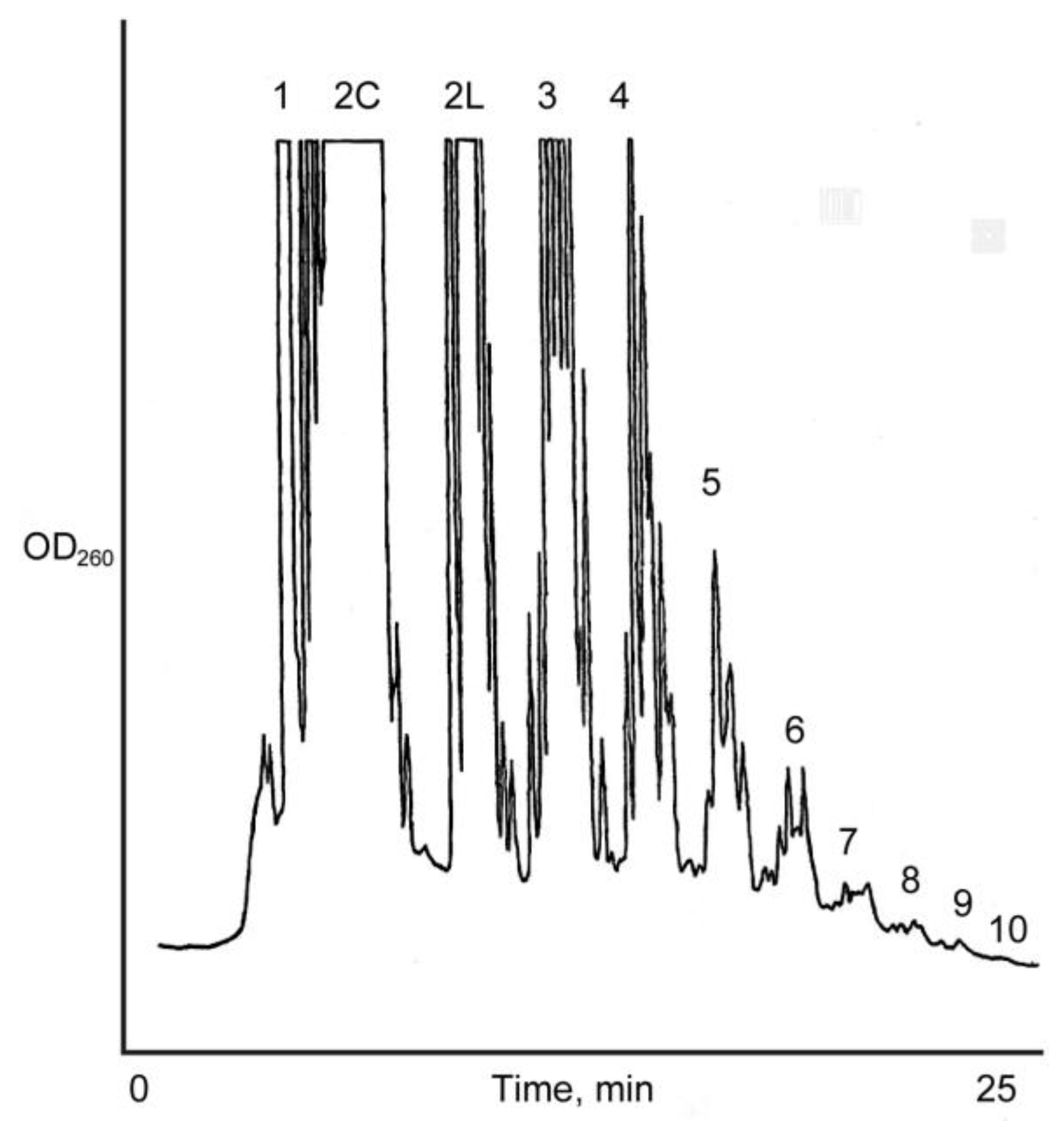
3.4. Effect of Salts in the Oligomerization of ImpA with Na+-Montmorillonite
| Reagent | Percentage Yield of Oligomers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | IIc * | IIL ** | III | IV | V | VI | VII | VIII | IX | X | XI | XII | |
| Effect of sodium chloride | |||||||||||||
| 2.0 M | 51.6 | 33.7 | 9.32 | 3.15 | 1.40 | 0.62 | 0.12 | 0.05 | 0.03 | T | |||
| 1.0 M | 24.9 | 52.2 | 13.6 | 4.84 | 2.29 | 1.12 | 0.58 | 0.27 | 0.14 | 0.05 | 0.01 | ||
| 0.2 M | 15.8 | 68.5 | 12.0 | 2.68 | 0.79 | 0.21 | 0.02 | T | |||||
| 0.1 M | 12.9 | 76.9 | 8.55 | 1.38 | 0.29 | 0.08 | |||||||
| Effect of sodium chloride (1 M) and/or magnesium chloride (0.075M) | |||||||||||||
| NaCl | 21.1 | 58.8 | 12.6 | 4.06 | 1.89 | 0.96 | 0.29 | 0.14 | 0.10 | 0.06 | T | ||
| MgCl2 | 22.0 | 57.4 | 12.7 | 4.70 | 1.83 | 0.80 | 0.35 | 0.13 | 0.06 | 0.03 | T | ||
| Effect of salt-in reagents(1 M) | |||||||||||||
| LiClO4 | 15.3 | 60.3 | 11.4 | 5.75 | 3.13 | 1.73 | 1.20 | 0.62 | 0.33 | 0.17 | 0.05 | 0.03 | |
| Guanidine | 53.2 | 26.1 | 10.5 | 4.58 | 2.39 | 1.41 | 0.81 | 0.47 | 0.26 | 0.17 | 0.05 | 0.04 | 0.02 |
| Effect of salt-out reagents(1 M) | |||||||||||||
| Na2SO4 | 11.4 | 60.5 | 13.0 | 6.17 | 3.36 | 1.93 | 1.40 | 0.81 | 0.52 | 0.40 | 0.32 | 0.19 | T |
| LiCl | 12.3 | 62.1 | 12.5 | 5.71 | 2.79 | 1.66 | 1.41 | 0.68 | 0.37 | 0.27 | 0.15 | 0.07 | T |
| Effect of reagents with no salt-in or salt-out effect(1 M) | |||||||||||||
| LiBr | 12.5 | 60.3 | 11.8 | 6.15 | 3.44 | 1.97 | 1.55 | 0.92 | 0.54 | 0.39 | 0.23 | 0.13 | 0.08 |
| Effect of no salts | |||||||||||||
| H2O | 34.5 | 63.7 | 1.80 | T | |||||||||
3.5. Effect of Hydrophilic and Hydrophobic Salts in the Oligomerization of ImpA Catalyzed by Na+-Montmorillonite
3.6. Effect of Monovalent Cations and Anions in the Oligomerization of ImpA Catalyzed by Na+-Montmorillonite
| Oligomer Length → | Percentage Yield of Oligomers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | IIc* | IIL** | III | IV | V | VI | VII | VIII | IX | X | XI | XII | |
| Effect of cations (1 M) with Chloride as a common ion | |||||||||||||
| LiCl | 12.3 | 62.0 | 12.5 | 5.68 | 2.79 | 1.65 | 1.41 | 0.68 | 0.36 | 0.29 | 0.22 | 0.12 | T |
| NaCl | 24.9 | 52.2 | 13.6 | 4.84 | 2.29 | 1.12 | 0.58 | 0.27 | 0.14 | 0.05 | 0.01 | T | |
| KCl | 72.2 | 21.2 | 5.29 | 1.10 | 0.34 | 0.05 | 0.02 | T | |||||
| Effect of anions (1M) with sodium as a common ion | |||||||||||||
| NaCl | 24.9 | 52.2 | 13.6 | 4.84 | 2.29 | 1.12 | 0.58 | 0.27 | 0.14 | 0.05 | 0.01 | T | |
| NaBr | 23.9 | 55.2 | 11.9 | 4.48 | 2.16 | 1.15 | 0.65 | 0.35 | 0.14 | 0.08 | T | ||
| NaI, 1M | 48.1 | 39.5 | 7.95 | 2.56 | 1.07 | 0.52 | 0.23 | 0.05 | 0.02 | T | |||
3.7. Effect of Homoionic Salts in the Oligomerization of ImpA Catalyzed by Na+-Montmorillonite
| Oligo Length→ Salt Concentration ↓ | Percentage Yield of Oligomers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | IIc* | IIL** | III | IV | V | VI | VII | VIII | IX | X | XI | |
| Li+-Montmorillonite | ||||||||||||
| LiCl, 1 M | 16.0 | 60.0 | 12.6 | 5.47 | 2.74 | 1.38 | 0.97 | 0.45 | 0.23 | 0.13 | 0.03 | T |
| NaCl, 1 M | 20.1 | 58.9 | 11.7 | 4.32 | 1.95 | 1.00 | 0.55 | 0.25 | 0.13 | 0.10 | T | |
| KCl, 1 M | 72.6 | 21.5 | 4.73 | 0.95 | 0.17 | 0.05 | T | |||||
| Na+-Montmorillonite | ||||||||||||
| LiCl, 1 M | 12.5 | 63.0 | 12.3 | 5.56 | 2.77 | 1.64 | 0.92 | 0.62 | 0.34 | 0.21 | 0.14 | T |
| NaCl, 1 M | 24.9 | 52.2 | 13.6 | 4.84 | 2.29 | 1.12 | 0.58 | 0.27 | 0.14 | 0.05 | 0.01 | T |
| KCl, 1 M | 72.0 | 21.2 | 5.29 | 1.10 | 0.35 | 0.04 | 0.02 | T | ||||
| K+-Montmorillonite | ||||||||||||
| LiCl, 1 M | 13.2 | 63.2 | 12.2 | 5.39 | 2.54 | 1.59 | 0.97 | 0.48 | 0.25 | 0.18 | ||
| NaCl, 1 M | 21.9 | 57.9 | 11.9 | 4.36 | 2.00 | 1.06 | 0.59 | 0.29 | T | |||
| KCl, 1 M | 73.4 | 21.0 | 4.54 | 0.92 | 0.12 | 0.02 | T | |||||
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cech, T.R.; Zung, A.J.; Grabowski, P.J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: Involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 1981, 27, 487–496. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Gilbert, W. The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Woese, C. (Ed.) The Genetic Code, the Molecular Basis for Genetic Expression; Harper and Row: New York, NY, USA, 1967.
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Orgel, L.E. A simple nucleic acid. Science 2000, 290, 1306–1307. [Google Scholar] [CrossRef]
- Szostak, J.W.; Ellington, A.D. In vivo Selection of Functional RNA Sequences. In The RNA World; Gesteland, R.K., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; pp. 511–533. [Google Scholar]
- Joyce, G.F.; Orgel, L.E. Prospects for Understanding the Origin of the RNA World. In The RNA World, 2nd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1999; pp. 49–77. [Google Scholar]
- Kazakov, S.A.; Altman, S. A trinucleotide can promote metal ion-dependent cleavage of RNA. Proc. Natl. Acad. Sci. USA 1992, 89, 7939–7943. [Google Scholar] [CrossRef]
- Turk, R.M.; Chumachenko, N.V.; Yarus, M. Multiple translational products from a five-nucleotide Ribozyme. Proc. Natl. Acad. Sci. USA 2010, 107, 4585–4589. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite catalysis of 30–50 mer oligonucleotides: Laboratory demonstration of potential steps in the origin of the RNA world. Orig. Life Evol. Biosph. 2002, 32, 311–332. [Google Scholar] [CrossRef]
- Huang, W.; Ferris, J.P. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc. 2006, 128, 8914–8919. [Google Scholar] [CrossRef]
- Joshi, P.C.; Pitsch, S.; Ferris, J.P. Chiral selectivity in the montmorillonite-catalyzed prebiotic synthesis of RNA. Chem. Commun. 2000, 24, 2497–2498. [Google Scholar] [CrossRef]
- Joshi, P.C.; Pitsch, S.; Ferris, J.P. Selectivity of montmorillonite catalyzed prebiotic reactions of d,l-nucleotides. Orig. Life Evol. Biosph. 2007, 37, 3–26. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Ferris, J.P. Progress in demonstrating total homochiral selection in montmorillonite-catalyzed RNA synthesis. Biochem. Biophys. Res. Commun. 2011, 413, 594–598. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Ferris, J.P. Homochiral selectivity in RNA synthesis: Montmorillonite-catalyzed quaternary reactions of d,l-purine with d,l-pyrimidine nucleotides. Orig. Life Evol. Biosph. 2011, 41, 213–236. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Ferris, J.P. Progress in Demonstrating Homochiral selection in Prebiotic RNA Synthesis. Adv. Space Res. 2013, 51, 772–779. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Delano, J.W.; Ferris, J.P. Mechanism of montmorillonite catalysis in the formation of RNA oligomers. J. Am. Chem. Soc. 2009, 131, 13369–13374. [Google Scholar] [CrossRef]
- Sleep, N.H. The hadean-archaena environment. Cold Spring Harb. Perspect. Biol. 2010, 2, a002527. [Google Scholar] [CrossRef]
- Knauth, L.P. Temperature and salinity history of the precambrian ocean: Implications for the course of microbial evolution. Palaegeogr. Palaeoclimatol. Palacoecol. 2005, 2190, 53–69. [Google Scholar] [CrossRef]
- Knauth, L.P. Salinity history of the Earth’s early ocean. Nature 1998, 395, 554–555. [Google Scholar] [CrossRef]
- Madsen, F.T.; Muller-Vonmoos, M. The swelling behavior of clays. Appl. Clay Sci. 1989, 4, 143–156. [Google Scholar] [CrossRef]
- Ponnuswamy, N.; Cougnon, F.B.L.; Clough, J.M.; Pantos, G.D.; Sanders, J.K.M. Discovery of an organic trefoil knot. Science 2012, 338, 783–785. [Google Scholar] [CrossRef]
- Kool, E.T.; Breslow, R. Dichotomous salt effects in the hydrophobic acceleration of benzoin condensation. J. Am. Chem. Soc. 1988, 110, 1596–1597. [Google Scholar] [CrossRef]
- Morrison, R.T.; Boyd, R.N. Organic Chemistry, 5th ed.; Allyn&Bacon, Inc.: Boston, MA, USA, 1987; p. 150. [Google Scholar]
- Sarfati, J. Origins of life: The chirality problem. J. Creat. 1998, 12, 263–266. [Google Scholar]
- Aldersley, M.F.; Joshi, P.C.; Price, J.D.; Ferris, J.P. The role of montmorillonite in its catalysis of RNA synthesis. Appl. Clay Sci. 2011, 54, 1–14. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F. Significance of mineral salts in Montmorillonite-catalyzed RNA synthesis. J. Mol. Evol. 2013, 76, 371–379. [Google Scholar] [CrossRef]
- Dickson, A.G.; Gayet, C. Handbook of Methods for the Analysis of the Various Parameters of the Carbon Dioxide System in Seawater, 2nd ed.; U.S. Department of Energy: Washington, DC, USA, 1994. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jheeta, S.; Joshi, P.C. Prebiotic RNA Synthesis by Montmorillonite Catalysis. Life 2014, 4, 318-330. https://doi.org/10.3390/life4030318
Jheeta S, Joshi PC. Prebiotic RNA Synthesis by Montmorillonite Catalysis. Life. 2014; 4(3):318-330. https://doi.org/10.3390/life4030318
Chicago/Turabian StyleJheeta, Sohan, and Prakash C. Joshi. 2014. "Prebiotic RNA Synthesis by Montmorillonite Catalysis" Life 4, no. 3: 318-330. https://doi.org/10.3390/life4030318
APA StyleJheeta, S., & Joshi, P. C. (2014). Prebiotic RNA Synthesis by Montmorillonite Catalysis. Life, 4(3), 318-330. https://doi.org/10.3390/life4030318




