Integrated Proteomic and Molecular Identification of Thermophilic Geobacillus Strains from Algerian Desert Sands and Their Enzymatic Potential
Abstract
1. Introduction
2. Materials and Methods
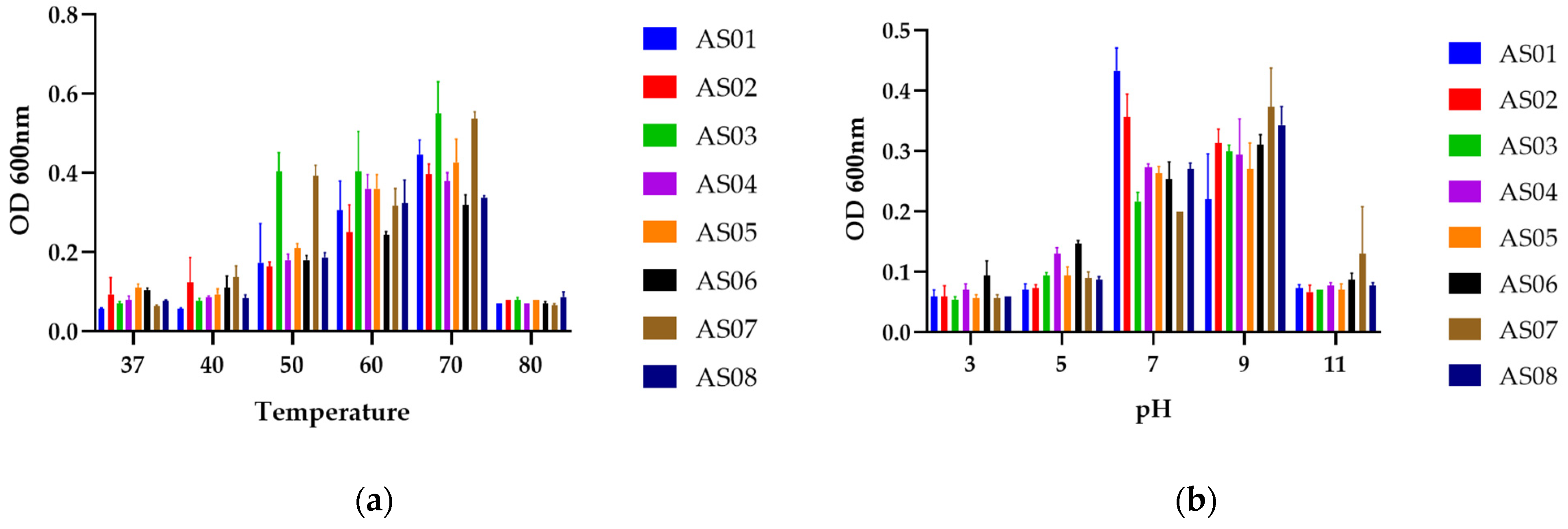
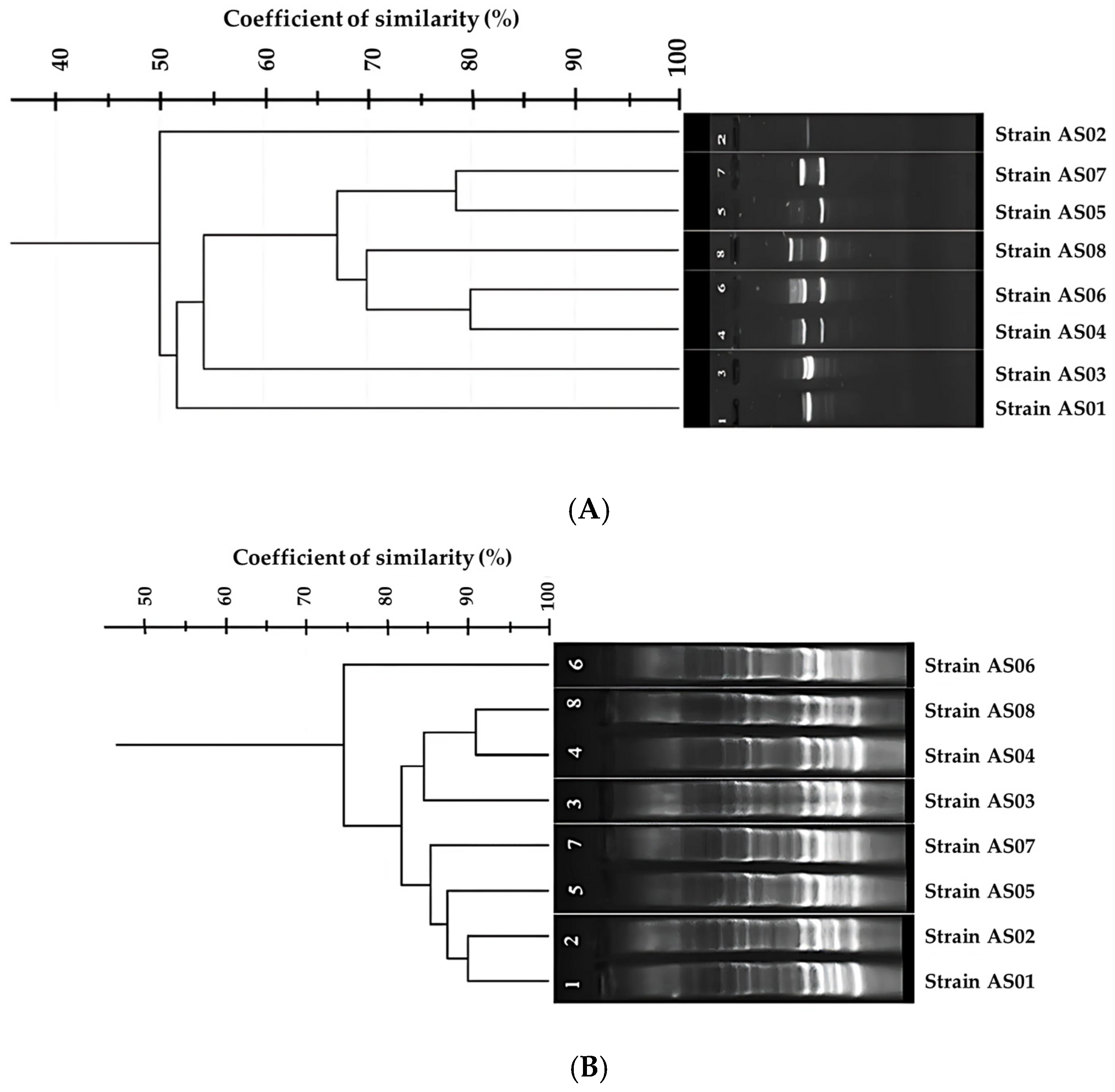
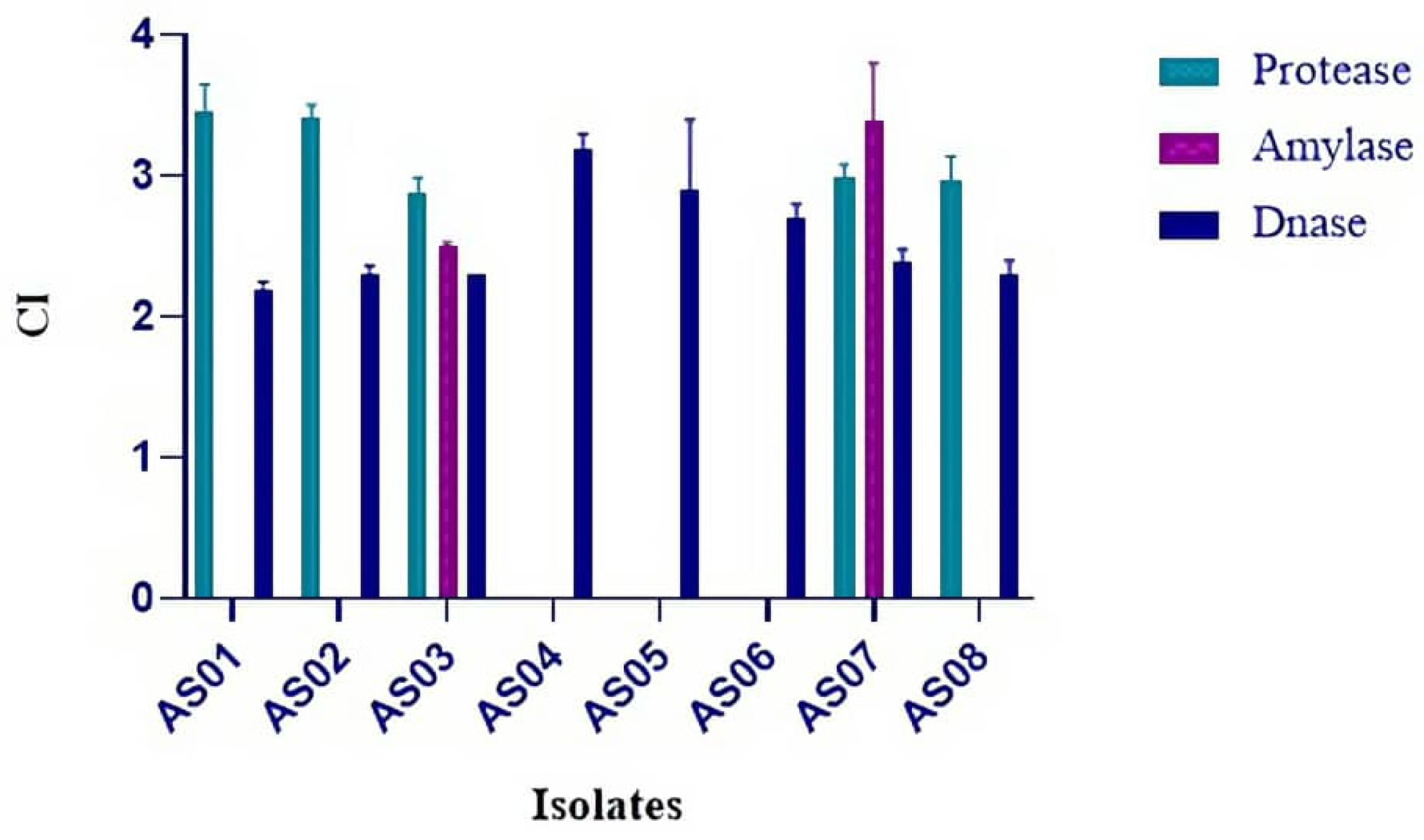
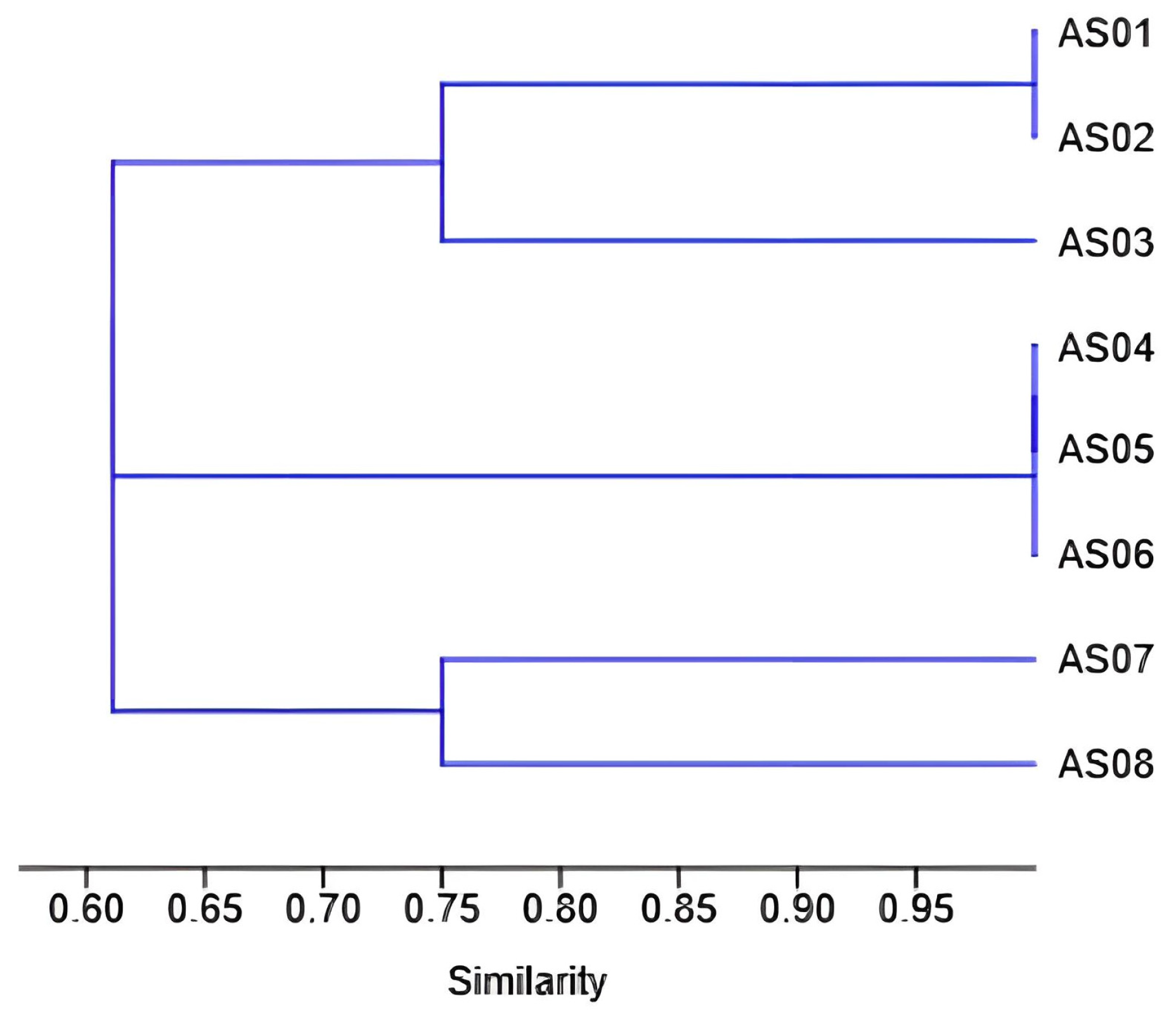
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valenzuela, B.; Solís-Cornejo, F.; Araya, R.; Zamorano, P. Isolation of Thermophilic Bacteria from Extreme Environments in Northern Chile. Microorganisms 2024, 12, 473. [Google Scholar] [CrossRef]
- Wang, Q.; Cen, Z.; Zhao, J. The survival mechanisms of thermophiles at high temperatures: An angle of omics. Physiology 2015, 30, 97–106. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Santana, M.M.; Gomez, E.J.; Delgado, J.A. Soil Thermophiles and Their Extracellular Enzymes: A Set of Capabilities Able to Provide Significant Services and Risks. Microorganisms 2023, 11, 1650. [Google Scholar] [CrossRef]
- Kumar, S.; Dangi, A.K.; Shukla, P.; Baishya, D.; Khare, S.K. Thermozymes: Adaptive strategies and tools for their biotechnological applications. Bioresour. Technol. 2019, 278, 372–382. [Google Scholar] [CrossRef]
- Gallo, G.; Imbimbo, P.; Aulitto, M. The Undeniable Potential of Thermophiles in Industrial Processes. Int. J. Mol. Sci. 2024, 25, 7685. [Google Scholar] [CrossRef]
- Khaswal, A.; Chaturvedi, N.; Mishra, S.K.; Kumar, P.R.; Paul, P.K. Current status and applications of genus Geobacillus in the production of industrially important products—A review. Folia Microbiol. 2022, 67, 389–404. [Google Scholar] [CrossRef] [PubMed]
- Kambourova, M. Thermostable enzymes and polysaccharides produced by thermophilic bacteria isolated from Bulgarian hot springs. Eng. Life Sci. 2018, 18, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; AdeelaYasid, N.; Shariff, F.M.; Abdul Rahman, N. Molecular Cloning, Characterization, and Application of Organic Solvent-Stable and Detergent-Compatible Thermostable Alkaline Protease from Geobacillus thermoglucosidasius SKF4. J. Microbiol. Biotechnol. 2024, 34, 436–456. [Google Scholar] [CrossRef] [PubMed]
- Baykara, S.G.; Sürmeli, Y.; Şanlı-Mohamed, G. Purification and Biochemical Characterization of a Novel Thermostable Serine Protease from Geobacillus sp. GS53. Appl. Biochem. Biotechnol. 2021, 193, 1574–1584. [Google Scholar] [CrossRef]
- Rigoldi, F.; Donini, S.; Redaelli, A.; Parisini, E.; Gautieri, A. Review: Engineering of thermostable enzymes for industrial applications. APL Bioeng. 2018, 2, 011501. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, F. Enzymes in Food Industry: Fermentation Process, Properties, Rational Design, and Applications. Foods 2024, 13, 3196. [Google Scholar] [CrossRef] [PubMed]
- Bibi, M.; Yasmin, A.; Blanford, C.; Safdar, N. Production and characterization of a highly stable laccase from extreme thermophile Geobacillus stearothermophilus MB600 isolated from hot spring of Gilgit Balitistan (Pakistan). J. Taibah Univ. Sci. 2023, 17, 2268903. [Google Scholar] [CrossRef]
- Jaiswal, N.; Jaiswal, P. Thermostable α-Amylases and Laccases: Paving the Way for Sustainable Industrial Applications. Processes 2024, 12, 1341. [Google Scholar] [CrossRef]
- Soy, S.; Nigam, V.K.; Sharma, S.R. Enhanced production and biochemical characterization of a thermostable amylase from thermophilic bacterium Geobacillus icigianus BITSNS038. J. Taibah Univ. Sci. 2021, 15, 730–745. [Google Scholar] [CrossRef]
- Lim, S.J.; Oslan, S.N. Native to designed: Microbial -amylases for industrial applications. PeerJ 2021, 9, e11315. [Google Scholar] [CrossRef]
- Bruins, M.E.; Janssen, A.E.; Boom, R.M. Thermozymes and their applications: A review of recent literature and patents. Appl. Biochem. Biotechnol. 2001, 90, 155–186. [Google Scholar] [CrossRef]
- Zainal Abidin, N.A.; Hasan, N.; Hamid, T.H.T.A. Isolation of thermophilic esterase or lipase producing bacteria from hot springs at the East coast of Peninsular Malaysia. J. Trop. Life Sci. 2021, 11, 1. [Google Scholar]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef]
- Kuisiene, N.; Raugalas, J.; Stuknyte, M.; Chitavichius, D. Identification of the genus Geobacillus using genus-specific primers, based on the 16S-23S rRNA gene internal transcribed spacer. FEMS Microbiol. Lett. 2007, 277, 165–172. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, S.; Park, Y.-S. Molecular typing tools for identifying and characterizing lactic acid bacteria: A review. Food Sci. Biotechnol. 2020, 29, 1301–1318. [Google Scholar] [CrossRef]
- Oubadi, M.; Faci, M. Spatio-Temporal Changes of Aridity in the Province of Naâma (Western Algeria). Ann. Arid. Zone 2024, 63, 41–49. [Google Scholar] [CrossRef]
- Ortega-Villar, R.; Escalante, A.; Astudillo-Melgar, F.; Lizárraga-Mendiola, L.; Vázquez-Rodríguez, G.A.; Hidalgo-Lara, M.E.; Coronel-Olivares, C. Isolation and Characterization of Thermophilic Bacteria from a Hot Spring in the State of Hidalgo, Mexico, and Geochemical Analysis of the Thermal Water. Microorganisms 2024, 12, 1066. [Google Scholar] [CrossRef]
- Sanwar Mal Yadav, R.P. Characterization of Endolithic Extremophilic Bacterial Diversity from Sandstone of Keroo, Jodhpur, Rajasthan. Int. J. Sci. Res. 2024, 13, 73–81. [Google Scholar] [CrossRef]
- Tripathi, N.; Zubair, M.; Sapra, A. Gram Staining. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Rafiee, Z.; Jalili Tabaii, M.; Moradi, M.; Harirchi, S. Unveiling Antibacterial Potential and Physiological Characteristics of Thermophilic Bacteria Isolated from a Hot Spring in Iran. Microorganisms 2024, 12, 834. [Google Scholar] [CrossRef]
- Li, Y.; Shan, M.; Zhu, Z.; Mao, X.; Yan, M.; Chen, Y.; Zhu, Q.; Li, H.; Gu, B. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect. Dis. 2019, 19, 941. [Google Scholar] [CrossRef]
- Adiguzel, A.; Ozkan, H.; Baris, O.; Inan, K.; Gulluce, M.; Sahin, F. Identification and characterization of thermophilic bacteria isolated from hot springs in Turkey. J. Microbiol. Methods 2009, 79, 321–328. [Google Scholar] [CrossRef]
- Ronimus, R.S.; Parker, L.E.; Morgan, H.W. The utilization of RAPD-PCR for identifying thermophilic and mesophilic Bacillus species. FEMS Microbiol. Lett. 1997, 147, 75–79. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Li, Y.; Liu, T.; Flint, S.; Zhang, G.; He, G. A RAPD based study revealing a previously unreported wide range of mesophilic and thermophilic spore formers associated with milk powders in China. Int. J. Food Microbiol. 2016, 217, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Gazali, F.M.; Suwastika, I.N. Thermostable α-Amylase Activity from Thermophilic Bacteria Isolated from Bora Hot Spring, Central Sulawesi. J. Phys. Conf. Ser. 2018, 979, 012001. [Google Scholar] [CrossRef]
- Sharma, K.; Pandit, S.; Thapa, B.S.; Pant, M. Biodegradation of Congo Red Using Co-Culture Anode Inoculum in a Microbial Fuel Cell. Catalysts 2022, 12, 1219. [Google Scholar] [CrossRef]
- Benammar, L.; İnan Bektaş, K.; Menasria, T.; Beldüz, A.O.; Güler, H.I.; Bedaida, I.K.; Gonzalez, J.M.; Ayachi, A. Diversity and enzymatic potential of thermophilic bacteria associated with terrestrial hot springs in Algeria. Braz. J. Microbiol. 2020, 51, 1987–2007. [Google Scholar] [CrossRef]
- Bouacem, K.; Bouanane-Darenfed, A. Isolation and characterization of thermophilic bacteria as producers of enzymes from Hammam Righa hot spring. In Proceedings of the MOL2NET′21, Conference on Molecular, Biomedical & Computational Sciences and Engineering, Basel, Switzerland, 25 January–30 December 2021; Volume 21, p. 12171. [Google Scholar]
- Aleem, B.; Rashid, M.H.; Zeb, N.; Saqib, A.; Ihsan, A.; Iqbal, M.; Ali, H. Random mutagenesis of super Koji (Aspergillus oryzae): Improvement in production and thermal stability of α-amylases for maltose syrup production. BMC Microbiol. 2018, 18, 200. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Singhal, P.; Singh, H.; Damle, D.; Sharma, A.K. Insight into thermophiles and their wide-spectrum applications. 3 Biotech 2016, 6, 81. [Google Scholar] [CrossRef]
- Arbab, S.; Ullah, H.; Khan, M.I.U.; Khattak, M.N.K.; Zhang, J.; Li, K.; Hassan, I.U. Diversity and distribution of thermophilic microorganisms and their applications in biotechnology. J. Basic Microbiol. 2022, 62, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Nuñez, L.F.; Rivera-Jacinto, M.A. Thermophilic bacteria from Peruvian hot springs with high potential application in environmental biotechnology. Environ. Technol. 2024, 45, 1420–1435. [Google Scholar] [CrossRef] [PubMed]
- Al-Mulla, A.; Khalifa, A. Isolation and Characterization of Thermophilic Bacteria Indigenous to Al-Ahsa Desert. J. Pure Appl. Microbiol. 2020, 14, 2157–2163. [Google Scholar] [CrossRef]
- Gomri, M.A.; El Moulouk Khaldi, T.; Kharroub, K. Analysis of the diversity of aerobic, thermophilic endospore-forming bacteria in two Algerian hot springs using cultural and non-cultural methods. Ann. Microbiol. 2018, 68, 915–929. [Google Scholar] [CrossRef]
- Adjeroud, M.; Escuder-Rodríguez, J.-J.; González-Siso, M.-I.; Kecha, M. Metagenomic Investiga-tion of Bacterial and Archaeal Diversity of Hammam Essalihine Hot Spring from Khenchela, Algeria. Geomicrobiol. J. 2020, 37, 804–817. [Google Scholar] [CrossRef]
- Turenne, C.Y.; Alexander, D.C. Bacillus and Other Aerobic Endospore-Forming Bacteria. In ClinMicroNow; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2023; pp. 1–21. ISBN 978-1-68367-043-8. [Google Scholar]
- Najar, I.N.; Sherpa, M.T.; Das, S.; Verma, K.; Dubey, V.K.; Thakur, N. Geobacillus yumthangensis sp. nov., a thermophilic bacterium isolated from a north-east Indian hot spring. Int. J. Syst. Evol. Microbiol. 2018, 68, 3430–3434. [Google Scholar] [CrossRef] [PubMed]
- Pawar, R.R.; Borkar, S.G. Isolation and characterization of thermophilic bacteria from different habitats and their assessment for antagonism against soil-borne fungal plant pathogens. Afr. J. Microbiol. Res. 2018, 12, 556–566. [Google Scholar] [CrossRef]
- Tang, R.; Yang, S.; Han, S.; Xie, C.-J.; Huang, G.-M.; Narsing Rao, M.P.; Liu, G.-H.; Zhou, S.-G. Bacillus litorisediminis sp. nov., a Thermophilic Bacterium Isolated from Mangrove Sediment. Curr. Microbiol. 2023, 80, 79. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Cortés, L.Y.; Ventura-Canseco, L.M.C.; Abud-Archila, M.; Ruíz-Valdiviezo, V.M.; Velázquez-Ríos, I.O.; Alvarez-Gutiérrez, P.E. Evaluation of temperature, pH and nutrient conditions in bacterial growth and extracellular hydrolytic activities of two Alicyclobacillus spp. strains. Arch. Microbiol. 2021, 203, 4557–4570. [Google Scholar] [CrossRef]
- O’Grady, J.; Cronin, U.; Tierney, J.; Piterina, A.V.; O’Meara, E.; Wilkinson, M.G. Chapter One-Gaps in the assortment of rapid assays for microorganisms of interest to the dairy industry. In Advances in Applied Microbiology; Gadd, G.M., Sariaslani, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 113, pp. 1–56. [Google Scholar]
- Zeigler, D.R. The Geobacillus paradox: Why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 2014, 160, 1–11. [Google Scholar] [CrossRef]
- Sharma, A.; Pandey, A.; Shouche, Y.S.; Kumar, B.; Kulkarni, G. Characterization and identification of Geobacillus spp. isolated from Soldhar hot spring site of Garhwal Himalaya, India. J. Basic Microbiol. 2009, 49, 187–194. [Google Scholar] [CrossRef]
- Yıldırım, İ.H.; Yıldırım, S.C.; Koçak, N. Molecular methods for bacterial genotyping and analyzed gene regions. J. Microbiol. Infect. Dis. 2011, 1, 42–46. [Google Scholar] [CrossRef][Green Version]
- Krader, P.; Emerson, D. Identification of archaea and some extremophilic bacteria using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Extremophiles 2004, 8, 259–268. [Google Scholar] [CrossRef]
- Tuohy, J.M.; Mueller-Spitz, S.R.; Albert, C.M.; Scholz-Ng, S.E.; Wall, M.E.; Noutsios, G.T.; Gutierrez, A.J.; Sandrin, T.R. MALDI-TOF MS Affords Discrimination of Deinococcus aquaticus Isolates Obtained from Diverse Biofilm Habitats. Front. Microbiol. 2018, 9, 2442. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef] [PubMed]
- Rychert, J. Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. J. Infect. Epidemiol. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- De Urraza, P.J.; Gómez-Zavaglia, A.; Lozano, M.E.; Romanowski, V.; De Antoni, G.L. DNA fingerprinting of thermophilic lactic acid bacteria using repetitive sequence-based polymerase chain reaction. J. Dairy Res. 2000, 67, 381–392. [Google Scholar] [CrossRef]
- Rai, P.; Sharma, A.; Saxena, P.; Soni, A.P.; Chakdar, H.; Kashyap, P.L.; Srivastava, A.K.; Sharma, A.K. Comparison of molecular and phenetic typing methods to assess diversity of selected members of the genus Bacillus. Microbiology 2015, 84, 236–246. [Google Scholar] [CrossRef]
- Yamamoto, K.; Murakami, R.; Takamura, Y. Differentiation of Thermophilic Anaerobic Gram-positive Bacteria by Random Amplified Polymorphic DNA Analysis. Microb. Environ. 2001, 16, 91–99. [Google Scholar] [CrossRef]
- Kwon, G.-H.; Lee, H.-A.; Park, J.-Y.; Kim, J.S.; Lim, J.; Park, C.-S.; Kwon, D.Y.; Kim, Y.-S.; Kim, J.H. Development of a RAPD-PCR method for identification of Bacillus species isolated from Cheonggukjang. Int. J. Food Microbiol. 2009, 129, 282–287. [Google Scholar] [CrossRef]
- Chuma, I.S.; Nonga, H.E.; Mdegela, R.H.; Kazwala, R.R. Epidemiology and RAPD-PCR typing of thermophilic campylobacters from children under five years and chickens in Morogoro Municipality, Tanzania. BMC Infect. Dis. 2016, 16, 692. [Google Scholar] [CrossRef]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef]
- Stefańska, I.; Kwiecień, E.; Górzyńska, M.; Sałamaszyńska-Guz, A.; Rzewuska, M. RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections. Pathogens 2022, 11, 562. [Google Scholar] [CrossRef]
- Moehling, T.J.; Browne, E.R.; Meagher, R.J. Effects of single and multiple nucleotide mutations on loop-mediated isothermal amplification. Analyst 2024, 149, 1701–1708. [Google Scholar] [CrossRef]
- Yoshida, R.; Nei, M. Efficiencies of the NJp, Maximum Likelihood, and Bayesian Methods of Phylogenetic Construction for Compositional and Noncompositional Genes. Mol. Biol. Evol. 2016, 33, 1618–1624. [Google Scholar] [CrossRef]
- Semenova, E.M.; Sokolova, D.S.; Grouzdev, D.S.; Poltaraus, A.B.; Vinokurova, N.G.; Tourova, T.P.; Nazina, T.N. Geobacillus proteiniphilus sp. nov., a thermophilic bacterium isolated from a high-temperature heavy oil reservoir in China. Int. J. Syst. Evol. Microbiol. 2019, 69, 3001–3008. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dietrich, C.H.; Wei, C. Genetic divergence, population differentiation and phylogeography of the cicada Subpsaltria yangi based on molecular and acoustic data: An example of the early stage of speciation? BMC Evol. Biol. 2019, 19, 5. [Google Scholar] [CrossRef]
- Burgess, S.A.; Flint, S.H.; Lindsay, D.; Cox, M.P.; Biggs, P.J. Insights into the Geobacillus stearothermophilus species based on phylogenomic principles. BMC Microbiol. 2017, 17, 140. [Google Scholar] [CrossRef]
- Lehmann, M.; Prohaska, C.; Zeldes, B.; Poehlein, A.; Daniel, R.; Basen, M. Adaptive laboratory evolution of a thermophile toward a reduced growth temperature optimum. Front. Microbiol. 2023, 14, 1265216. [Google Scholar] [CrossRef]
- Leng, H.; Wang, Y.; Zhao, W.; Sievert, S.M.; Xiao, X. Identification of a deep-branching thermophilic clade sheds light on early bacterial evolution. Nat. Commun. 2023, 14, 4354. [Google Scholar] [CrossRef]
- Abdul Majid, S.; Graw, M.F.; Chatziefthimiou, A.D.; Nguyen, H.; Richer, R.; Louge, M.; Sultan, A.A.; Schloss, P.; Hay, A.G. Microbial Characterization of Qatari Barchan Sand Dunes. PLoS ONE 2016, 11, e0161836. [Google Scholar] [CrossRef]
- Ulucay, O.; Gormez, A.; Ozic, C. Identification, characterization and hydrolase producing performance of thermophilic bacteria: Geothermal hot springs in the Eastern and Southeastern Anatolia Regions of Turkey. Antonie Van Leeuwenhoek 2022, 115, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Thebti, W.; Riahi, Y.; Gharsalli, R.; Belhadj, O. Screening and characterization of thermo-active enzymes of biotechnological interest produced by thermophilic Bacillus isolated from hot springs in Tunisia. Acta Biochim. Pol. 2016, 63, 581–587. [Google Scholar] [CrossRef]
- Anand, A.; Kumar, V.; Satyanarayana, T. Characteristics of thermostable endoxylanase and β-xylosidase of the extremely thermophilic bacterium Geobacillus thermodenitrificans TSAA1 and its applicability in generating xylooligosaccharides and xylose from agro-residues. Extremophiles 2013, 17, 357–366. [Google Scholar] [CrossRef]
- Valenzuela, B.; Solís-Cornejo, F.; Araya, R.; Zamorano, P. Isolation and Characterization of Thermus thermophilus Strain ET-1: An Extremely Thermophilic Bacterium with Extracellular Thermostable Proteolytic Activity Isolated from El Tatio Geothermal Field, Antofagasta, Chile. Int. J. Mol. Sci. 2023, 24, 14512. [Google Scholar] [CrossRef]
- Ward, O.P.; Moo-Young, M. Thermostable enzymes. Biotechnol. Adv. 1988, 6, 39–69. [Google Scholar] [CrossRef]
- Gomes, E.; de Souza, A.R.; Orjuela, G.L.; Da Silva, R.; de Oliveira, T.B.; Rodrigues, A. Applications and Benefits of Thermophilic Microorganisms and Their Enzymes for Industrial Biotechnology. In Gene Expression Systems in Fungi: Advancements and Applications; Schmoll, M., Dattenböck, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 459–492. ISBN 978-3-319-27951-0. [Google Scholar]
- Li, J.; Sun, L.; Huo, Y.-X. High-Temperature Catalytic Platform Powered by Thermophilic Microorganisms and Thermozymes. Synth. Biol. Eng. 2025, 3, 10001. [Google Scholar] [CrossRef]
- Pham, T.V.; Hua, T.C.; Nguyen, N.A.; Nguyen, H.T.D. Purification and Characterization of a Small Thermostable Protease from Streptomyces sp. CNXK100. Pol. J. Microbiol. 2024, 73, 155–165. [Google Scholar] [CrossRef]
- DeCastro, M.-E.; Rodríguez-Belmonte, E.; González-Siso, M.-I. Metagenomics of Thermophiles with a Focus on Discovery of Novel Thermozymes. Front. Microbiol. 2016, 7, 1521. [Google Scholar] [CrossRef]
- Kumar, A.; Bhanja Dey, T.; Mishra, A.K.; Meena, K.R.; Mohapatra, H.S.; Kuhad, R.C. Optimization and Characterization of an Ultra-Thermostable, Acidophilic, Cellulase-Free Xylanase from a New Obligate Thermophilic Geobacillus thermoleovorans AKNT10 and Its Application in Saccharification of Wheat Bran. Curr. Microbiol. 2024, 81, 287. [Google Scholar] [CrossRef]
- Bhalla, A.; Arce, J.; Ubanwa, B.; Singh, G.; Sani, R.K.; Balan, V. Thermophilic Geobacillus WSUCF1 Secretome for Saccharification of Ammonia Fiber Expansion and Extractive Ammonia Pretreated Corn Stover. Front. Microbiol. 2022, 13, 844287. [Google Scholar] [CrossRef]
- Khadka, S.; Khadka, D.; Poudel, R.C.; Bhandari, M.; Baidya, P.; Sijapati, J.; Maharjan, J. Production Optimization and Biochemical Characterization of Cellulase from Geobacillus sp. KP43 Isolated from Hot Spring Water of Nepal. Biomed Res. Int. 2022, 2022, 6840409. [Google Scholar] [CrossRef]
- Li, H.; Hu, Q.; Hong, X.; Jiang, Z.; Ni, H.; Li, Q.; Zhu, Y. Molecular Cloning and Characterization of a Thermostable and Halotolerant Endo-β-1,4-Glucanase from Microbulbifer sp. ALW1. 3 Biotech 2021, 11, 250. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, J.; Wang, J.; Lin, L.; Wei, W.; Shen, Y.; Wei, D. A Type I Pullulanase from Geobacillus subterraneus: Functional Expression in Escherichia coli, Enzyme Characterization, Truncation, and Application. Starch-Stärke 2022, 74, 2200044. [Google Scholar] [CrossRef]



| Isolates Code | Detected Species | MALDI-TOF Score |
|---|---|---|
| AS01 | Geobacillus kaustophilus | 1.91 |
| AS02 | Geobacillus kaustophilus | 2.25 |
| AS03 | Geobacillus jurassicus | 1.86 |
| AS04 | Geobacillus kaustophilus | 1.98 |
| AS05 | Geobacillus kaustophilus | 2.02 |
| AS06 | Geobacillus kaustophilus | 2.14 |
| AS07 | Geobacillus jurassicus | 1.98 |
| AS08 | Geobacillus kaustophilus | 2.13 |
| Strain | Closest Species | % Similarity | Accession Number |
|---|---|---|---|
| AS01 | Geobacillus proteiniphilus strain 1017 | 99.46% | NR_178413.1 |
| AS02 | Geobacillus kaustophilus NBRC 102445 | 99.03% | NR_114089.1 |
| AS03 | Geobacillus kaustophilus NBRC 102445 | 99.87% | NR_114089.1 |
| AS04 | Geobacillus thermoleovorans strain LEH-1 | 97.98% | NR_036985.1 |
| AS05 | Geobacillus proteiniphilus strain 1017 | 98.26% | NR_178413.1 |
| AS06 | Geobacillus kaustophilus NBRC 102445 | 100% | NR_114089.1 |
| AS07 | Geobacillus kaustophilus NBRC 102445 | 98.91% | NR_114089.1 |
| AS08 | Geobacillus thermoleovorans strain LEH-1 | 99.86% | NR_036985.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammadi, A.I.; Merzoug, M.; Aireche, M.; Zater, Z.Y.; Bendida, K.; Brakna, C.N.; Choubane, S.; Todorov, S.D.; Saidi, D. Integrated Proteomic and Molecular Identification of Thermophilic Geobacillus Strains from Algerian Desert Sands and Their Enzymatic Potential. Life 2025, 15, 1327. https://doi.org/10.3390/life15081327
Hammadi AI, Merzoug M, Aireche M, Zater ZY, Bendida K, Brakna CN, Choubane S, Todorov SD, Saidi D. Integrated Proteomic and Molecular Identification of Thermophilic Geobacillus Strains from Algerian Desert Sands and Their Enzymatic Potential. Life. 2025; 15(8):1327. https://doi.org/10.3390/life15081327
Chicago/Turabian StyleHammadi, Amaria Ilhem, Mohamed Merzoug, Marwa Aireche, Zohra Yasmine Zater, Keltoum Bendida, Chaimaa Naila Brakna, Slimane Choubane, Svetoslav Dimitrov Todorov, and Djamal Saidi. 2025. "Integrated Proteomic and Molecular Identification of Thermophilic Geobacillus Strains from Algerian Desert Sands and Their Enzymatic Potential" Life 15, no. 8: 1327. https://doi.org/10.3390/life15081327
APA StyleHammadi, A. I., Merzoug, M., Aireche, M., Zater, Z. Y., Bendida, K., Brakna, C. N., Choubane, S., Todorov, S. D., & Saidi, D. (2025). Integrated Proteomic and Molecular Identification of Thermophilic Geobacillus Strains from Algerian Desert Sands and Their Enzymatic Potential. Life, 15(8), 1327. https://doi.org/10.3390/life15081327







