Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Propagation, Inoculation with Virus, RNA Extraction, and RT-PCR
2.2. Constructs Prepared in the Current Study
2.3. Transformation of Agrobacterium tumefaciens and Generation of Transgenic A. thaliana Plants
2.4. Total Plant Protein Extraction, SDS-PolyAcrylamide Gel Electrophoresis (PAGE), and Western Blot Analysis
2.5. Methodology for Observation of Morphological Symptoms of A. thaliana Plants in Response to Virus/Viral Component Infection
2.6. Bioinformatic Analysis
2.6.1. Library Construction and Sequencing by LC Sequencing Company
2.6.2. Uses of Bioinformatics Software in Identifying circRNAs Relevant to the Current Study
2.7. circRNA Classification Based on Genomic Origins
3. Results Obtained
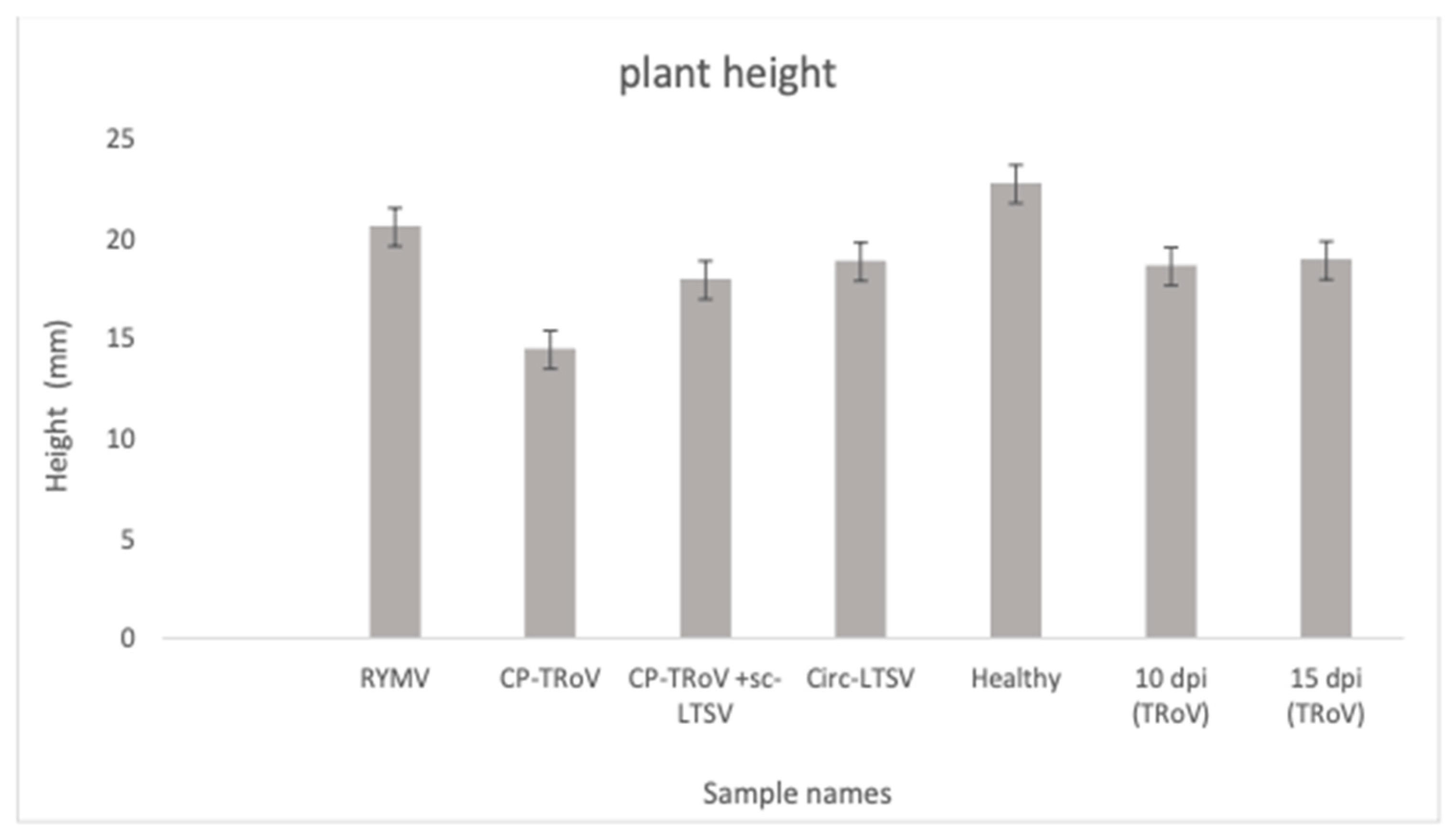
3.1. Symptom Development in A. thaliana Plants Due to Viral Infection
ANOVA and Tukey HSD Statistical Analyses
3.2. Methodological Confirmations of Clones and Transgenic Plants Used in the Current Study
3.2.1. Detection of CP-TRoV, RYMV, and circ-LTSV Genes in Transgenic A. thaliana Plants
Presence of CP-TRoV Gene in Transgenic CP-TRoV A. thaliana
Presence of Genomic RYMV Transgene in RYMV Transgenic A. thaliana
Confirmation of scLTSV in Transgenic A. thaliana
Confirmation of Circular scLTSV (circ-LTSV) Replication in Transgenic A. thaliana Infected with TRoV
Confirmation of the Absence of CP-TRoV in scLTSV-Transgenic A. thaliana
3.2.2. Western Blot Analysis
Western Blot Analysis for the Detection of CP Expression in Plants Transgenic for CP-TRoV
Absence of RYMV Capsid Protein Expression in Transgenic A. thaliana as Demonstrated by Western Blot Analysis
3.3. Major Findings from the Current Study
3.3.1. CircRNA Sequencing and Profiling
3.3.2. Results Obtained from RNA-SEQ Bioinformatic Analyses: Identification and Characterization of circRNAs, and Analysis of circRNA Abundance Across Samples and GC Content
3.3.3. Genomic Distribution of Identified circRNAs
3.3.4. Principal Component Analysis
3.3.5. Chromosomal Distribution of circRNAs
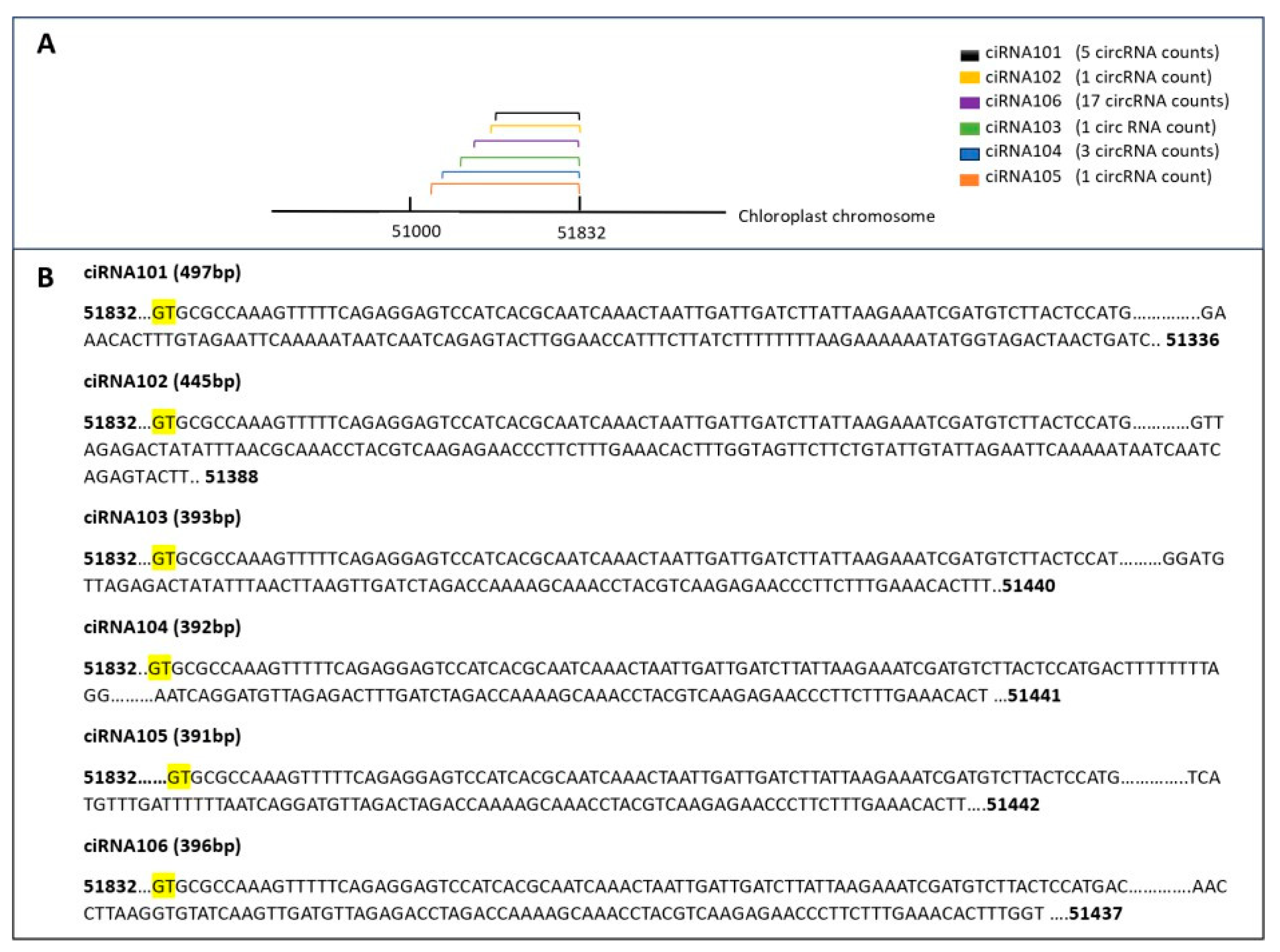
3.3.6. Diversity in circRNA Splicing Signals
3.3.7. Patterns of circRNA Counts
3.3.8. Gene Ontology (GO) Functional Analysis
3.3.9. Characteristics of Chloroplast-Derived circRNAs
3.3.10. Internal ORF Identification in circRNAs
3.3.11. Functional Insights of Identified circRNAs
3.3.12. CircRNA–miRNA Interactions
3.3.13. Enrichment Analysis
3.3.14. Validation of Identified circRNAs by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santer, L.; Bär, C.; Thum, T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol. Ther. 2019, 27, 1350–1363. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Darbani, B.; Noeparvar, S.; Borg, S. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front. Plant Sci. 2016, 7, 776. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Sun, X.; Liu, Y.; Li, H.; Deng, G.; Lin, H.; Wang, S. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol. Biol. 2018, 96, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Zhang, H.; Wang, H.; Liu, X.; Xu, X.; Zhang, Z.; Kohnen, M.V.; Hu, K.; Wang, H.; et al. Genome-Wide profiling of circular RNAs in the rapidly growing shoots of moso bamboo (Phyllostachys edulis). Plant Cell. Physiol. 2019, 60, 1354–1373. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Izadpanah, K.; Peters, J.R.; Dietzgen, R.G.; Mitter, N. Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci. 2018, 274, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Gao, L.; Zhu, B.; Luo, Y.; Deng, Z.; Zuo, J. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol. Plant 2017, 161, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Jia, J.H.; Yang, L.; Xia, Y.C.; Zhang, H.L.; Jia, J.B.; Zhou, R.; Nie, P.Y.; Yin, J.L.; Ma, D.F.; et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H.; Fan, M.; He, Y.; Guo, P. Genome-wide identification of long non-coding RNAs and circular RNAs reveal their ceRNA networks in response to cucumber green mottle mosaic virus infection in watermelon. Arch. Virol. 2020, 165, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Antoniw, J.F. DPV–Database of Plant Viruses. Available online: https://www.dpvweb.net/ (accessed on 17 July 2025).
- Broadbent, L.; Heathcote, G.D. Properties and host range of turnip crinkle, rosette and yellow mosaic viruses. Ann. Appl. Biol. 1958, 46, 585. [Google Scholar] [CrossRef]
- Sehgal, O.P.; Sinha, R.C.; Gellatly, D.L.; Ivanov, I.; AbouHaidar, M.G. Replication and encapsidation of the viroid-like satellite RNA of lucerne transient streak virus are supported in divergent hosts by cocksfoot mottle virus and turnip rosette virus. J. Gen. Virol. 1993, 74, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.47658 (accessed on 17 July 2025).
- Philips, A.; Nowis, K.; Stelmaszczuk, M.; Jackowiak, P.; Podkowiński, J.; Handschuh, L.; Figlerowicz, M. Expression Landscape of circRNAs in Arabidopsis thaliana Seedlings and Adult Tissues. Front. Plant Sci. 2020, 11, 576581. [Google Scholar] [CrossRef] [PubMed]
- Callaway, A.S.; George, C.G.; Lommel, S.A. A Sobemovirus coat protein gene complements long-distance movement of a coat protein-null Dianthovirus. Virology 2004, 330, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Nummert, G.; Sõmera, M.; Uffert, G.; Abner, E.; Truve, E. P1-independent replication and local movement of Rice yellow mottle virus in host and non-host plant species. Virology 2017, 502, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Abouhaidar, M.G.; Paliwal, Y.C. Comparison of the nucleotide sequences of viroid-like satellite RNA of the Canadian and Australasian strains of lucerne transient streak virus. J. Gen. Virol. 1988, 69, 2369–2373. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Wen-Sheng, X.; Xiang-Jing, W.; Tian-Rui, R.; Su-Qin, C. Purification of recombinant wheat cytochrome P450 monooxygenase expressed in yeast and its properties. Protein Expr. Purif. 2006, 45, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, M.; Nishikiori, M.; Tomita, K.; Yoshioka, N.; Kozuka, R.; Naito, S.; Ishikawa, M. The Arabidopsis Cucumovirus Multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004, 78, 6102–6111. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Ohta, M.; Takahashi, T.; Meshi, R.; Schmidt, C.; Dean, S.; Naito, M.; Ishikawa, M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 2000, 97, 10107–10112. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Park, K.Y.; Seo, Y.S.; Kim, W.T. Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 2016, 170, 2494–2510. [Google Scholar] [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Zhang, P.; Berardini, T. Using the Arabidopsis Information Resource (TAIR) to Find Information About Arabidopsis Genes. Curr. Protoc. 2022, 2, e574. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Bai, P.; Zhu, X.; Zhang, X.; Mao, L.; Zhu, Q.H.; Fan, L.; Ye, C.Y. Characteristics of plant circular RNAs. Brief. Bioinform. 2020, 21, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Zhang, H.; Wang, J.; Zinta, G.; Xie, S.; Zhu, W.; Nie, W.F. Genome-Wide Identification of Circular RNAs in Response to Low-Temperature Stress in Tomato Leaves. Front. Genet. 2020, 11, 591806. [Google Scholar] [CrossRef] [PubMed]
- Yie, Y.; Tien, P. Plant virus satellite RNAs and their role in engineering resistance to virus diseases. Semin. Virol. 1993, 4, 363–368. [Google Scholar] [CrossRef]
- Lin, K.Y.; Lin, N.S. Interfering Satellite RNAs of Bamboo mosaic virus. Front. Microbiol. 2017, 8, 787. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of Circular RNAs and Their Targets in Leaves of Triticum aestivum L. under Dehydration Stress. Front. Plant Sci. 2017, 7, 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.; Guo, B.; Xu, L.; Li, L.; Song, X.; Wang, X.; Zeng, X.; Wu, L.; Niu, D.; et al. Exonic Circular RNAs Are Involved in Arabidopsis Immune Response Against Bacterial and Fungal Pathogens and Function Synergistically with Corresponding Linear RNAs. Phytopathology 2022, 112, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Hébrard, E.; Poulicard, N.; Gérard, C.; Traoré, O.; Wu, H.C.; Albar, L.; Fargette, D.; Bessin, Y.; Vignols, F. Direct Interaction Between the Rice yellow mottle virus (RYMV) VPg and the Central Domain of the Rice eIF(iso)4G1 Factor Correlates with Rice Susceptibility and RYMV Virulence. Mol. Plant Microb. Interact. 2010, 23, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Sanfaçon, H. Plant translation factors and virus resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Abudraham, S.; Shaya, F.; Ostersetzer-Biran, O. An optimized method for the analysis of plant mitochondria RNAs by Northern-blotting. Endocytobiosis Cell Res. 2011, 21, 34–42. [Google Scholar]
















| Primer Name | Sequence (5′-3′) |
|---|---|
| TRoV CP-F | GAGGACCAATTCAGTGGTTACACC |
| TRoV CP-R | CTGCTGCCGTTGTTCCATCAGCGG |
| LTSV_sat_F | CCTACCATGGCCTCATCAGT |
| LTSV_sat_R | GCCGGTAGGATGATGGATTA |
| circ-LTSV-F | GGTCGACTCTAGAGGATCCCCCCCATGGCCTCATCAGT |
| circ-LTSV-R | GGCGCGGTCTAGATACGACTCACTATAGGGCGAATTCG |
| RYMV-F | ACAATTGAAG CTAGGAAAGG AGC |
| RYMV-R | CTCCCCCACC CATCCCGAGA ATT |
| Analysis Item | Software | Version |
|---|---|---|
| Quality control | FastQC | 0.10.1 |
| Adapter removal | Cutadapt | 1.10 |
| Genome mapping | Tophat | 2.0.4 |
| Back-splicing junction reads filter | Tophat-fusion | 2.1 |
| circRNA identification | CIRCexplore, CIRI | 2.2.6, 2.0.2 |
| Differential expression analysis | edgeR | NA |
| Interaction with miRNA | Targetmimics | NA |
| CircRNA ID | Reverse Primer | Forward Primer | Product Size |
|---|---|---|---|
| circRNA2 | TGATTCCACTTCCTTCGATGC | ACTGTAATCGCGGCTTTGTC | 142 bp |
| ciRNA33 | TGAGCACATTGAACTATCCATGT | TACTCCTCCTGTTCGAACCG | 115 bp |
| ciRNA65 | CCTAATGTCAGGCTGTTGTTCT | TGATCTTTTCGTCCTATGAACCT | 167 bp |
| ciRNA6 | CCTAATGTCAGGCTGTTGTTCT | AGCAAACCTACGTCAAGAGAAC | 135 bp |
| circRNA205 | ACTCCCAAGCGCACAAATTC | TTTCTTCTTAGCGGCTTGGC | 195 bp |
| ciRNA9 | TGCTAATGTGCCTTGGATGA | TGCGTTCGGGAAGGATGAAT | 125 bp |
| CircRNA105 | GTAAAAGCAAGATGATACTTC | GGTGACACAAGGATTTTCAG | 182 bp |
| ciRNA91 | TCAGGCTGTTGTTCTCCTCTT | ACCAAAAGCAAACCTACGTCA | 196 bp |
| ciRNA17 | TGCTAATGTGCCTTGGATGA | TGCGTTCGGGAAGGATGAAT | 188 bp |
| ciRNA3 | ATTCGCGGGATTATGCACTC | GCATGAGAGCCAAATGAATCG | 134 bp |
| circRNA296 | TAGTACTTGCCTAGCGGACG | TGGTGGAGATTCTAGGCGAC | 187 bp |
| circRNA248 | TCCCTATCGAGTCAAAAGGAAGA | GTTCAGAACCGGATGATCAAGA | 158 bp |
| circRNA61 | TATGCCTGCTTTGACCGAGA | TGCAAGATCACGTCCCTCAT | 157 bp |
| circRNA247 | TGCACATAAGGCGTTTCATTT | GTGGCTATGGACGTTGCTAA | 125 bp |
| CircRNA221 | CCTTTCGGAGCTTCAACAGT | CTGGGATGATGAGGATGTGGA | 165 bp |
| ciRNA8 | ACTCCCTGCATTTAATTCCACT | GCGACTTGTCAGATATATCGGG | 200 bp |
| Elongation factor alpha housekeeping gene (EF) | |||
| Primer ID | Reverse primer | Forward primer | Product size |
| CC2012 (qRT-PCR) | GGTCTGCCTCATGTCCCTAA | TGGTGACGCTGGTATGGTTA | 109 bp |
| EI382 (RT-PCR) | GCTCCTGGTCATCGTGATTT | GCTCCTGGTCATCGTGATTT | 488 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashim, G.M.; Haight, T.; Chen, X.; Zovoilis, A.; Venkataraman, S. Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection. Life 2025, 15, 1143. https://doi.org/10.3390/life15071143
Hashim GM, Haight T, Chen X, Zovoilis A, Venkataraman S. Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection. Life. 2025; 15(7):1143. https://doi.org/10.3390/life15071143
Chicago/Turabian StyleHashim, Ghyda Murad, Travis Haight, Xinyang Chen, Athanasios Zovoilis, and Srividhya Venkataraman. 2025. "Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection" Life 15, no. 7: 1143. https://doi.org/10.3390/life15071143
APA StyleHashim, G. M., Haight, T., Chen, X., Zovoilis, A., & Venkataraman, S. (2025). Profiling Plant circRNAs Provides Insights into the Expression of Plant Genes Involved in Viral Infection. Life, 15(7), 1143. https://doi.org/10.3390/life15071143





