Isolation, Sphalerite Bioleaching, and Whole Genome Sequencing of Acidithiobacillus ferriphilus QBS3 from Zinc-Rich Sulfide Mine Drainage
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description, Sampling, and Isolation of Bioleaching Bacteria
2.2. Identification of the Bioleaching Strain
2.3. Growth, Oxidation, and Bioleaching Study
2.4. Whole Genome Sequencing, Assembly, and Annotation
2.5. Evolutionary Analyses and Functional Evaluation of Bioleaching-Related Genes
2.6. Average Nucleotide Identity (ANI) and DDH Calculations
2.7. Pan-Genome Analyses of A. ferriphilus spp.
3. Results
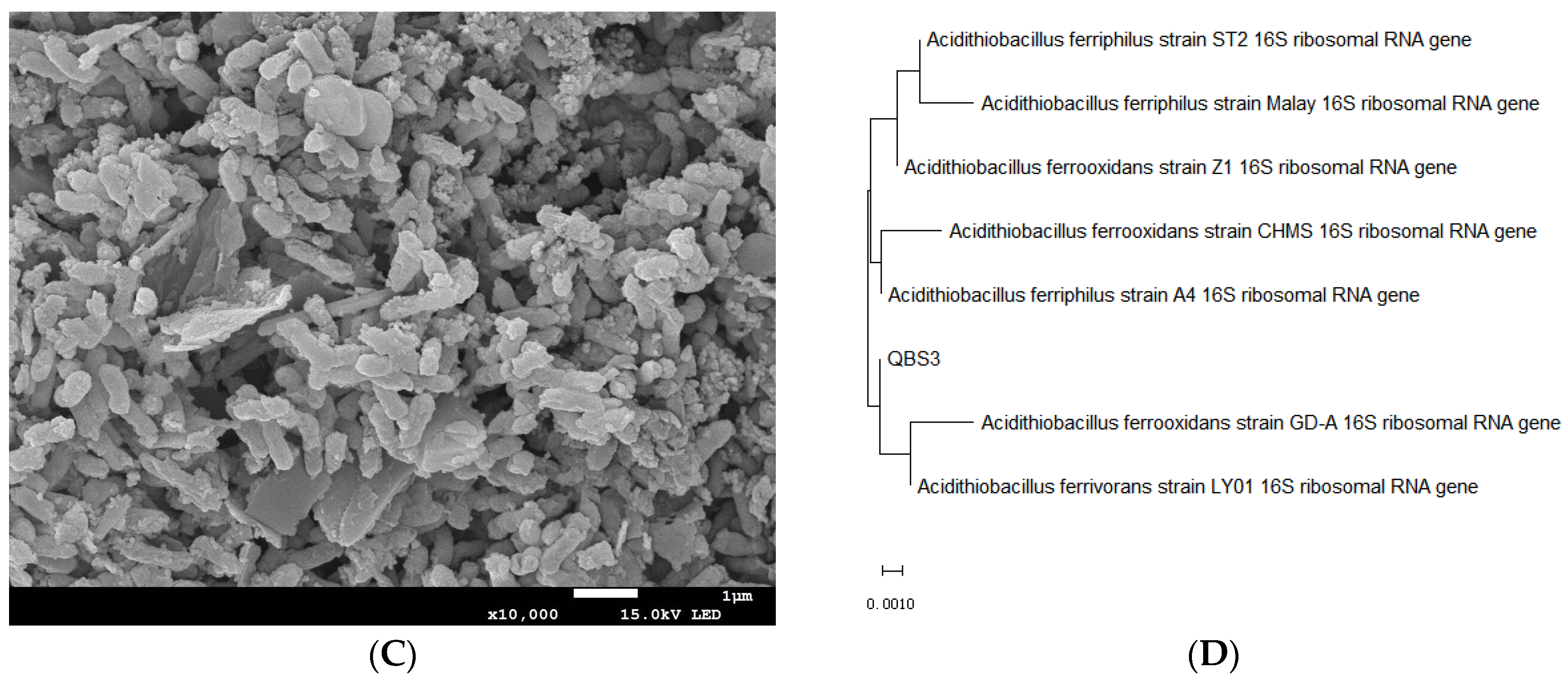
3.1. Isolation and Characterization of Bioleaching Bacteria
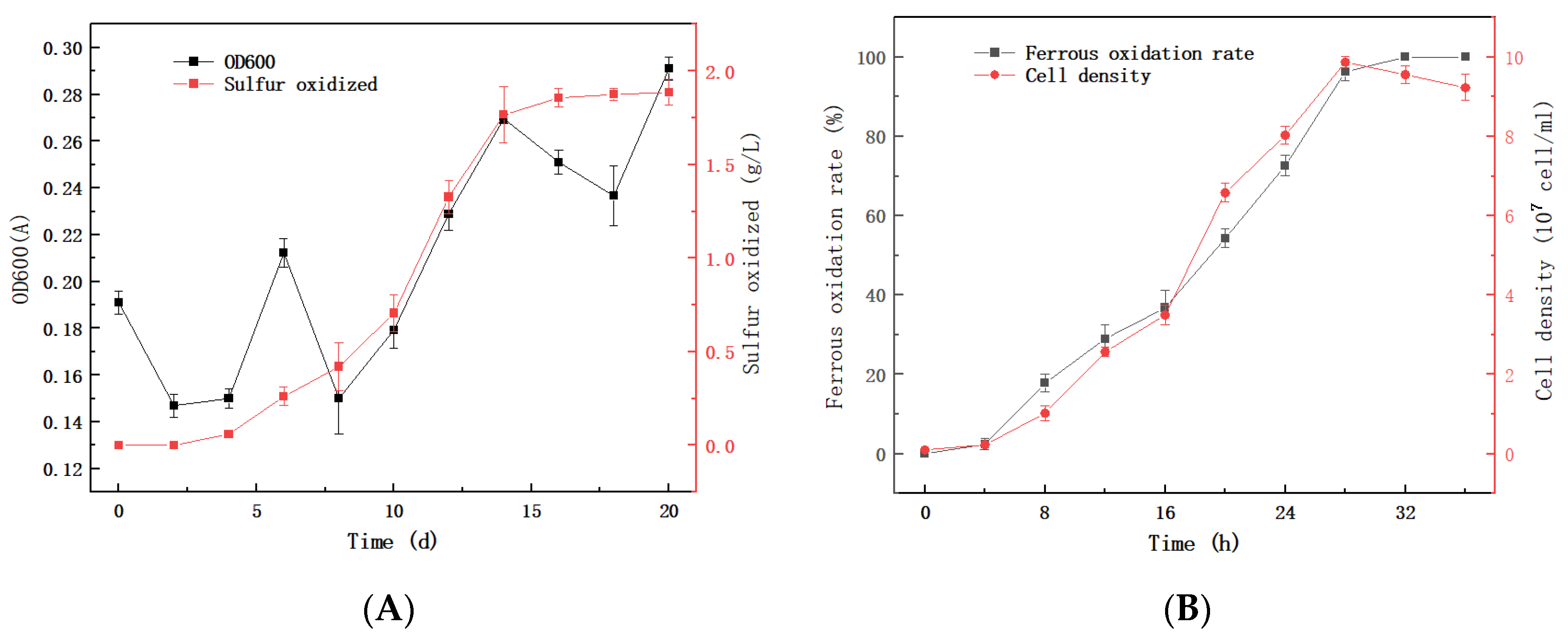
3.2. Physiological and Biochemical Characterization of QBS3
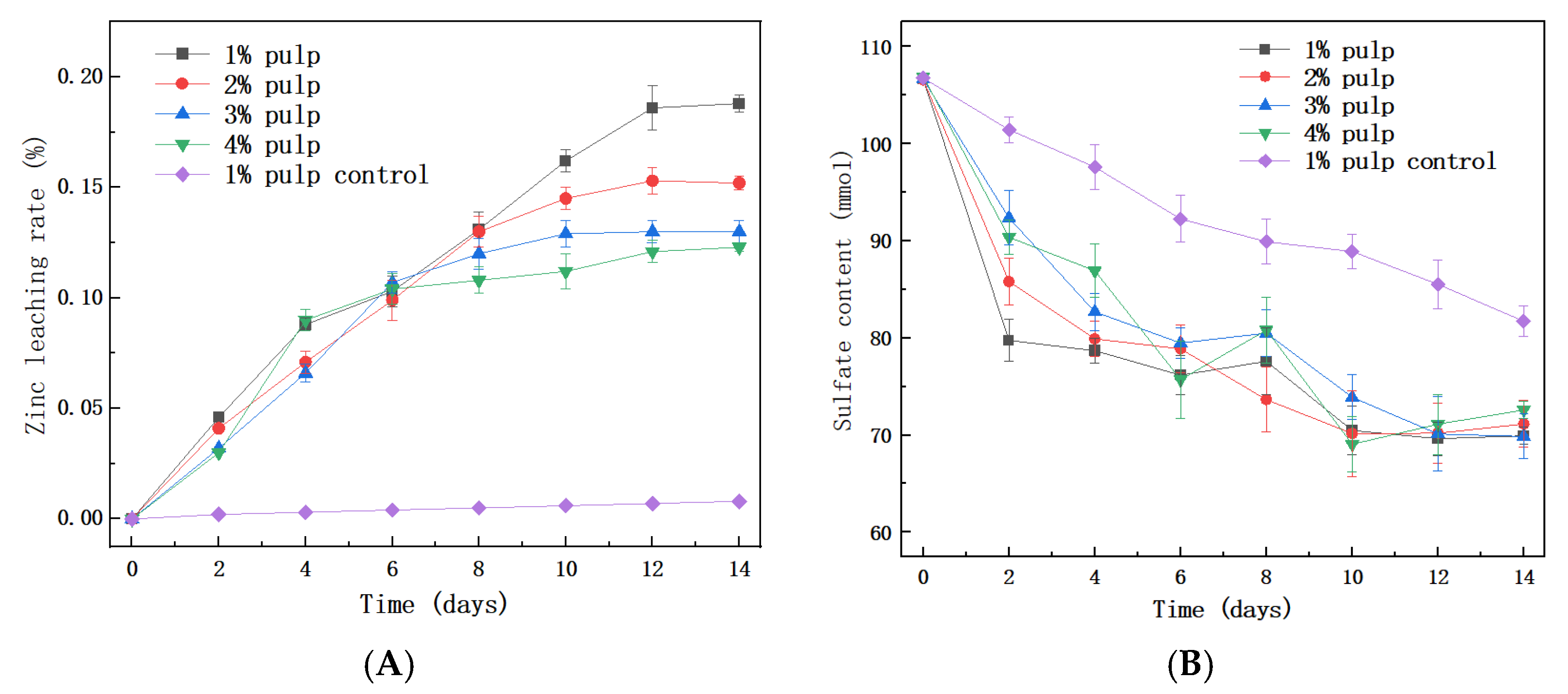
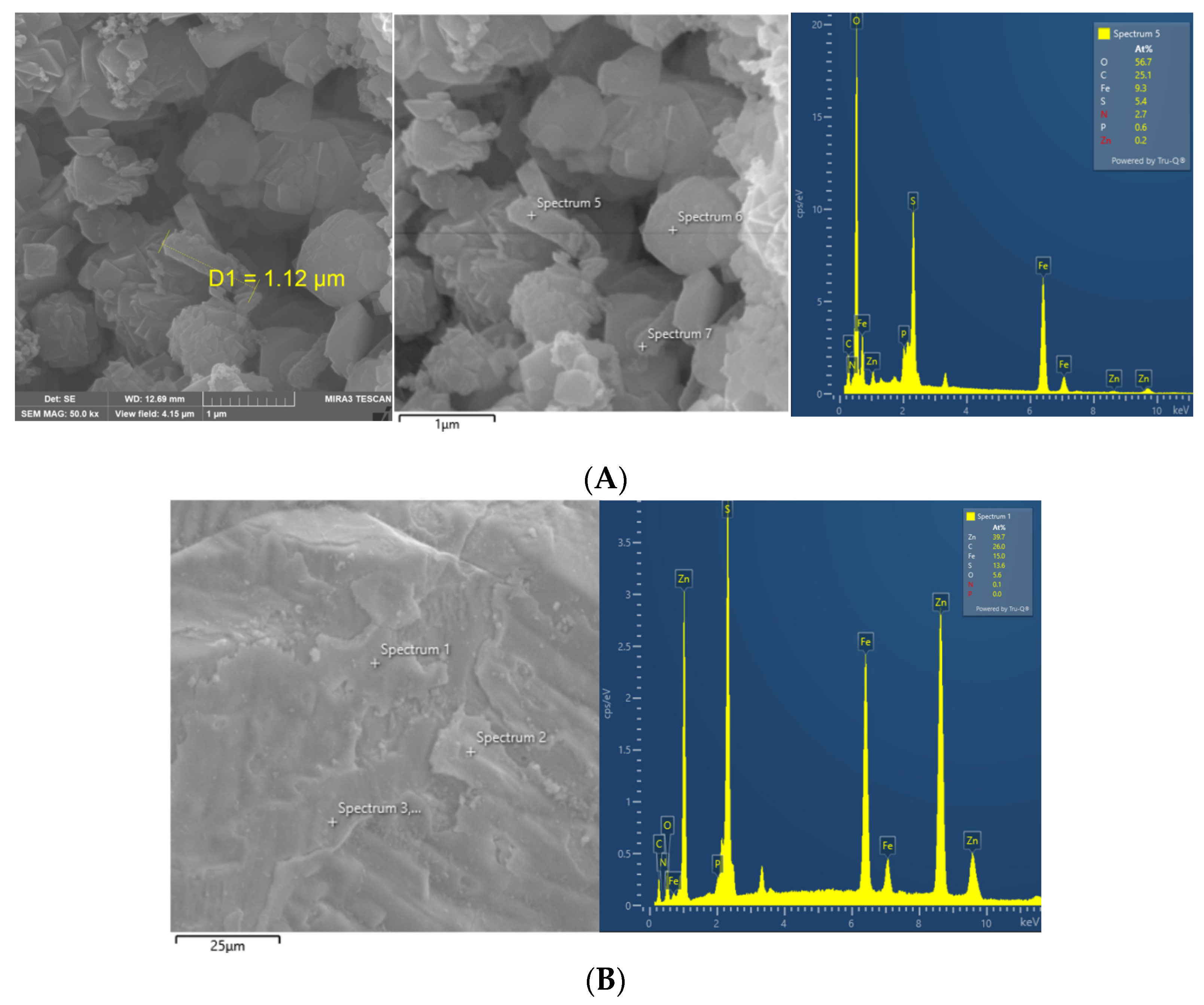
3.3. The Sphalerite Bioleaching Effect of QBS3
3.4. Genomic Characteristics of QBS3
3.5. Phylogenetic Analyses of QBS3
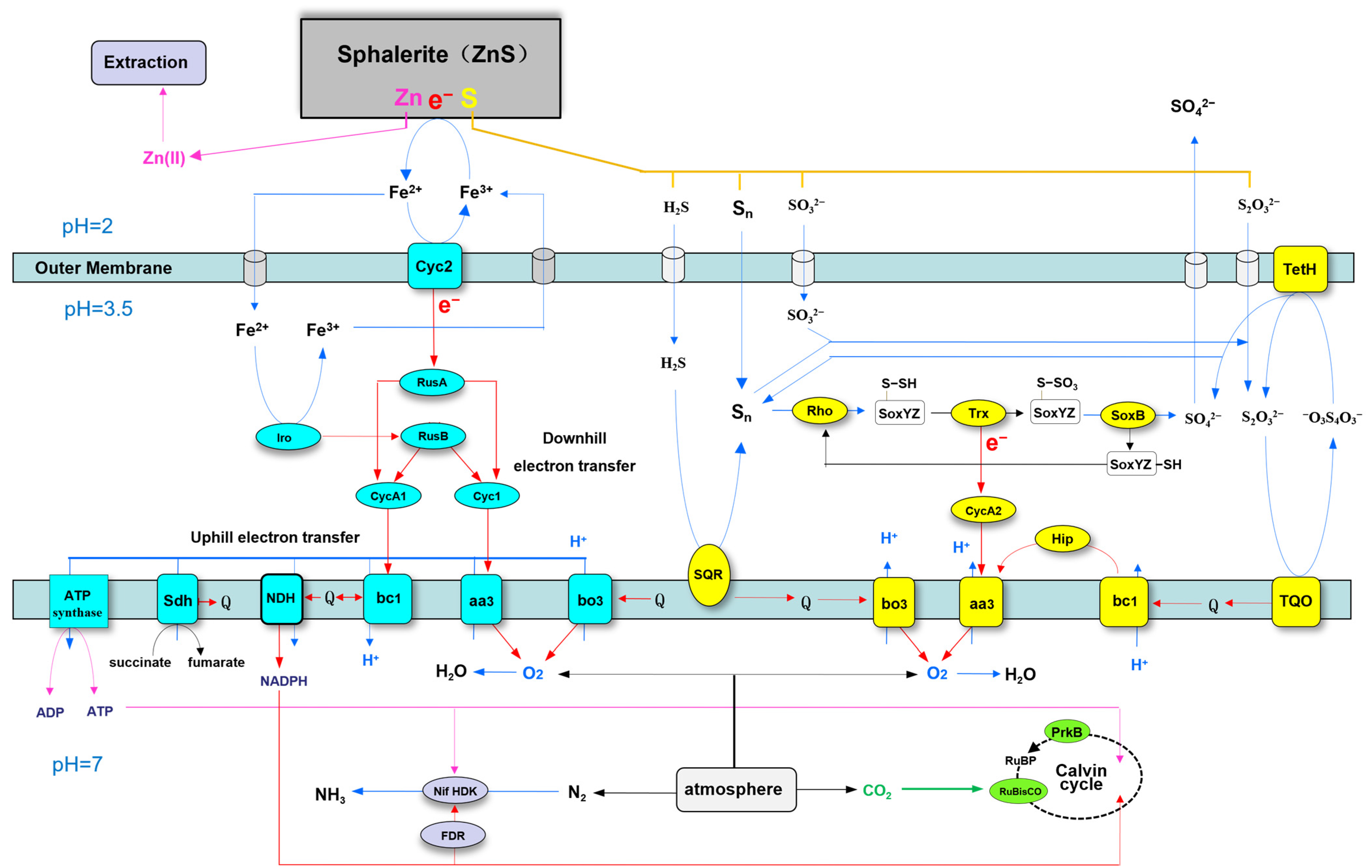
3.6. Sphalerite Bioleaching-Related Genes of A. ferriphilus QBS3
3.6.1. Ferrous Oxidation-Related Genes
3.6.2. Sulfur Oxidation-Related Genes
3.6.3. Respiratory Chain Complex-Related Genes
3.6.4. Carbon and Nitrogen Fixation-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| XRD | X-ray Diffraction |
| XRF | X-ray Fluorescence |
| ICP | Inductively Coupled Plasma |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
References
- Shi, S.; Fang, Z.; Ni, J. Comparative study on the bioleaching of zinc sulphides. Process Biochem. 2006, 41, 438–446. [Google Scholar] [CrossRef]
- Syurin, S.; Vinnikov, D. Occupational disease predictors in the nickel pyrometallurgical production: A prospective cohort observation. J. Occup. Med. Toxicol. 2022, 17, 21. [Google Scholar] [CrossRef]
- Jones, S.; Santini, J.M. Mechanisms of bioleaching: Iron and sulfur oxidation by acidophilic microorganisms. Essays Biochem. 2023, 67, 685–699. [Google Scholar] [CrossRef]
- Ubaldini, S.; Veglió, F.; Toro, L.; Abbruzzese, C. Biooxidation of arsenopyrite to improve gold cyanidation: Study of some parameters and comparison with grinding. Int. J. Miner. Process. 1997, 52, 65–80. [Google Scholar] [CrossRef]
- Zou, G.; Papirio, S.; Lai, X.; Wu, Z.; Zou, L.; Puhakka, J.; Ruan, R. Column leaching of low-grade sulfide ore from Zijinshan copper mine. Int. J. Miner. Process. 2015, 139, 11–16. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R.; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genom. 2008, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Falagán, C.; Johnson, D.B. Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 2016, 66, 206–211. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, A.; Yang, Y.; Liu, J.; Qiu, G. Isolation, characterization of Acidiphilium sp. DX1-1 and ore bioleaching by this acidophilic mixotrophic organism. Trans. Nonferrous Met. Soc. China 2013, 23, 1774–1782. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Amanze, C.; Zheng, X.; Man, M.; Yu, Z.; Ai, C.; Wu, X.; Xiao, S.; Xia, M.; Yu, R.; Wu, X.; et al. Recovery of heavy metals from industrial wastewater using bioelectrochemical system inoculated with novel Castellaniella species. Environ. Res. 2022, 205, 112467. [Google Scholar] [CrossRef]
- Tabrizi, A.B. Development of a dispersive liquid–liquid microextraction method for iron speciation and determination in different water samples. J. Hazard. Mater. 2010, 183, 688–693. [Google Scholar] [CrossRef]
- Pina, P.S.; Leão, V.A.; Silva, C.A.; Daman, D.; Frenay, J. The effect of ferrous and ferric iron on sphalerite bioleaching with Acidithiobacillus sp. Miner. Eng. 2005, 18, 549–551. [Google Scholar] [CrossRef]
- Brown, C.G.; Clarke, J. Nanopore development at Oxford Nanopore. Nat. Biotechnol. 2016, 34, 810–811. [Google Scholar] [CrossRef]
- Pak, T.R.; Sullivan, M.; Attie, O.; Webster, E.; Kasarskis, A.; van Bakel, H.; Bashir, A. PathogenDB: A Modular Software Suite Integrating Genomic Clinical Microbiology and Epidemiology. Open Forum Infect. Dis. 2016, 3, 1479. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Bertelli, C.; Brinkman, F.S.L.; Valencia, A. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007, 8, 209. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucl. Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan-genome analysis web service. Imeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Singh, A.; Lahiri, C.; Deb, C.; Roy, P. Colony morphology mutants of chemolithotrophic Acidithiobacillus ferrooxidans are associated with altered genomic distribution of family 1 repetitive DNA sequence. Curr. Sci. Assoc. 2002, 82, 1009–1014. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Tsaplina, I.A.; Kondrat’eva, T.F.; Belyi, A.V.; Bulaev, A.G. Physiological and Morphological Characteristics of Acidophilic Bacteria Leptospirillum ferriphilum and Acidithiobacillus thiooxidans, Members of a Chemolithotrophic Microbial Consortium. Microbiology 2018, 87, 326–338. [Google Scholar] [CrossRef]
- Liu, h.; Wang, X.; Long, J.; Gao, Y.; Wan, Z.; Liu, C. Effect of Electric Field on Culture of Acidithiobacillus Ferrooxidans. Nonferrous Met. Extr. Metall. 2022, 8, 76–81. [Google Scholar]
- Li, W.; Feng, Q.; Li, Z. Isolation and Characterization of A Novel Iron–Sulfur Oxidizing Bacterium Acidithiobacillus Ferrooxidans YQ-N3 and its Applicability in Coal Biodesulfurization. Minerals 2023, 13, 95. [Google Scholar] [CrossRef]
- Bhaskar, S.; Manoj, A.; Nayak, D.M.; Furtado, I.M.; Anchan, D.; Wazir, A.M. Influence of Ferrous Iron Addition on Silver Catalyzed Bioleaching of Copper from Chalcopyrite using an Isolated Acidithiobacillus ferrooxidans Strain. Grenze Int. J. Eng. Technol. 2023, 2. [Google Scholar] [CrossRef]
- Abdollahi, H.; Mirmohammadi, M.; Ghassa, S.; Jozanikohan, G.; Boroumand, Z.; Tuovinen, O.H. Acid bioleaching of select sphalerite samples of variable Zn- and Fe-contents. Hydrometallurgy 2022, 212, 105897. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, X.; Zhang, L.; Zheng, H.; Dong, Z.; Gu, G.; Shen, L.; Zhao, H. Bioleaching of sphalerite mineralized with discrepant iron incorporations: Experiment and DFT calculation. J. Mater. Res. Technol. 2022, 19, 794–808. [Google Scholar] [CrossRef]
- Khaleque, H.N.; Kaksonen, A.H.; Boxall, N.J.; Watkin, E.L.J. Chloride ion tolerance and pyrite bioleaching capabilities of pure and mixed halotolerant, acidophilic iron- and sulfur-oxidizing cultures. Miner. Eng. 2018, 120, 87–93. [Google Scholar] [CrossRef]
- Lan, Z.; Li, D.; Zhang, Q. Electrochemical Study on the Bioleaching of Marmatite. Adv. Mater. Res. 2013, 774–776, 512–518. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Zhang, L.; Tang, A.; Lv, X.; Zhao, Y.; Shen, L.; Zhao, H.; Qiu, G. Effects of Mechanical Activation on the Bioleaching of Sphalerite and Marmatite for Zn Extraction. Minerals 2021, 11, 111. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Jiang, V.; Khare, S.D.; Banta, S. Computational Structure Prediction Provides a Plausible Mechanism for Electron Transfer by the Outer Membrane Protein Cyc2 from Acidithiobacillus ferrooxidans. Protein Sci. 2021, 30, 1640–1652. [Google Scholar] [CrossRef]
- Liu, Y.; He, J.; Shangguan, X.; Liu, R.; Zeng, X.; Belqadi, W.; Wang, K.; Tong, Y.; Yu, R.; Zeng, W.; et al. Purification, Characterization, and Ferrous Oxidation Kinetics of Iron Oxidase from Acidithiobacillus ferridurans. Separations 2023, 10, 554. [Google Scholar] [CrossRef]
- Sasaki, K.; Ida, C.; Ando, A.; Matsumoto, N.; Saiki, H.; Ohmura, N. Respiratory Isozyme, Two Types of Rusticyanin of Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 2003, 67, 1039–1047. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, Z.; Jiang, Y.; Dong, Y.; Shi, L. In vivo interactions between Cyc2 and Rus as well as Rus and Cyc1 of Acidithiobacillus ferrooxidans during extracellular oxidization of ferrous iron. Int. Biodeterior. Biodegrad. 2022, 173, 105453. [Google Scholar] [CrossRef]
- Zhang, Y.; Weiner, J.H. Characterization of the kinetics and electron paramagnetic resonance spectroscopic properties of Acidithiobacillus ferrooxidans sulfide:quinone oxidoreductase (SQR). Arch. Biochem. Biophys. 2014, 564, 110–119. [Google Scholar] [CrossRef]
- Cwalina, B.; Wilczok, T.; Dzierżewicz, Z.; Weglarz, L. Activity of sulphite oxidase, thiosulphate oxidase and rhodanese in Thiobacillus ferrooxidans during sphalerite leaching. Acta Microbiol. Pol. 1990, 39, 99–103. [Google Scholar]
- Wang, Y.; Zhang, X.; Liu, Q.; Ai, C.; Mo, H.; Zeng, J. Expression, Purification and Molecular Structure Modeling of Thioredoxin (Trx) and Thioredoxin Reductase (TrxR) from Acidithiobacillus ferrooxidans. Curr. Microbiol. 2009, 59, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Kanao, T.; Sharmin, S.; Tokuhisa, M.; Otsuki, M.; Kamimura, K. Identification of a gene encoding a novel thiosulfate:quinone oxidoreductase in marine Acidithiobacillus sp. strain SH. Res. Microbiol. 2020, 171, 281–286. [Google Scholar] [CrossRef]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y. Extending the models for iron and sulfur oxidation in the extreme Acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009, 10, 394. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Y.; Yu, R. Differential of Transcription Expression&Bioinformatics Tapping of a Cytochrome c Family Gene of afe_0378 in Acidithiobacillus ferrooxidans. J. Microbiol. 2014, 34, 7–11. [Google Scholar]
- Calisto, F.; Todorovic, S.; Louro, R.O.; Pereira, M.M. Exploring substrate interaction in respiratory alternative complex III from Rhodothermus marinus. BBA Bioenerg. 2023, 1864, 148983. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Yang, M.; Zhang, S.; Zhao, D.; Duan, J.; Wang, W.; Yan, L. Iron and sulfur oxidation pathways of Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 2019, 35, 60. [Google Scholar] [CrossRef]
- Esparza, M.; Cárdenas, J.P.; Bowien, B.; Jedlicki, E.; Holmes, D.S. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiol. 2010, 10, 229–243. [Google Scholar] [CrossRef]
- Amouric, A.; Brochier-Armanet, C.; Johnson, D.B.; Bonnefoy, V.; Hallberg, K.B. Phylogenetic and genetic variation among Fe(II)- oxidizing acidithiobacilli supports the view that these comprise multiple species with different ferrous iron oxidation pathways(Article). Microbiology 2011, 157, 111–122. [Google Scholar] [CrossRef]
- Patra, M.C.; Pradhan, S.K.; Rath, S.N.; Maharana, J. Structural Analysis of Respirasomes in Electron Transfer Pathway of Acidithiobacillus ferrooxidans: A Computer-Aided Molecular Designing Study. ISRN Biophys. 2013, 2013, 295718. [Google Scholar] [CrossRef]
- Landry, A.P.; Ballou, D.P.; Banerjee, R. Hydrogen Sulfide Oxidation by Sulfide Quinone Oxidoreductase. Chembiochem 2021, 22, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Welte, C.; Hafner, S.; Krätzer, C.; Quentmeier, A.; Friedrich, C.G.; Dahl, C. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 2009, 583, 1281–1286. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Rother, D.; Carius, Y. The structure of the periplasmic thiol-disulfide oxidoreductase SoxS from Paracoccus pantotrophus indicates a triple Trx/Grx/DsbC functionality in chemotrophic sulfur oxidation. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 229–240. [Google Scholar] [CrossRef]
- Peketi, A.; Mazumdar, A.; Pyne, P.; Ghosh, W.; Alam, M. Kinetic Enrichment of S-34 during Proteobacterial Thiosulfate Oxidation and the Conserved Role of SoxB in S-S Bond Breaking. Appl. Environ. Microbiol. 2013, 79, 4455–4464. [Google Scholar] [CrossRef]
- Ibáñez, A.; Garrido-Chamorro, S.; Coque, J.J.R.; Barreiro, C. From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus. Genes 2023, 14, 1772. [Google Scholar] [CrossRef] [PubMed]
- Kanao, T.; Hase, N.; Nakayama, H.; Yoshida, K.; Nishiura, K.; Kosaka, M.; Kamimura, K.; Hirano, Y.; Tamad, T. Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Protein Sci. 2021, 30, 328–338. [Google Scholar] [CrossRef]
- Taha, T.M.; Takeuchi, F.; Sugio, T. Reduction of Cytochrome c by Tetrathionate in the Presence of Tetrathionate Hydrolase Purified from Sulfur-Grown Acidithiobacillus Ferrooxidans ATCC 23270. Adv. Mater. Res. 2009, 71–73, 243–246. [Google Scholar] [CrossRef]
- Qin, X. Study on the Genes Involved in the Ferrous Oxidation and Sulfur Oxidation in Acidithiobacillus ferrooxidans. Ph.D. Thesis, Shandong University, Jinan, China, 2018. [Google Scholar]



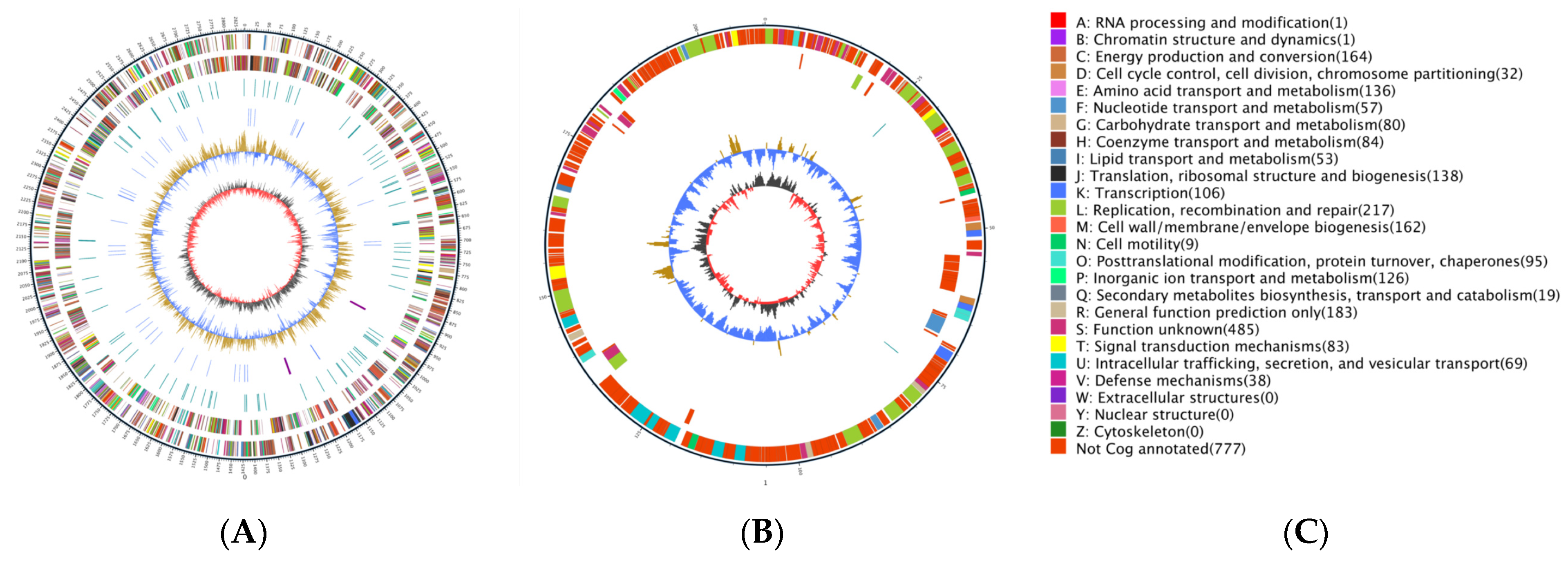


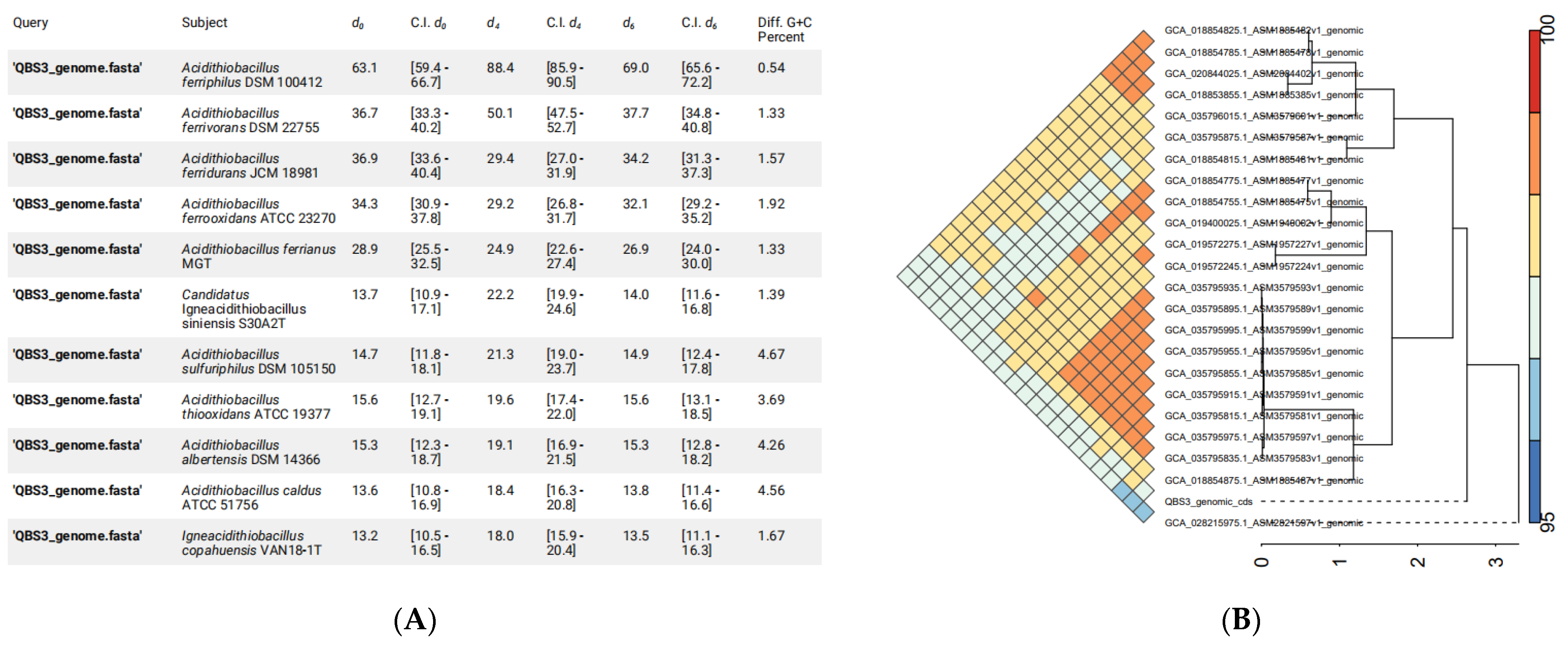

| Name | Location | Start | End | Length |
|---|---|---|---|---|
| Genomic_island_1 | Chromosome | 737,433 | 746,842 | 9410 |
| Genomic_island_2 | Chromosome | 1,424,882 | 1,438,714 | 13,833 |
| Genomic_island_3 | Chromosome | 1,606,329 | 1,621,443 | 15,115 |
| Genomic_island_4 | Chromosome | 1,662,644 | 1,676,150 | 13,507 |
| Genomic_island_5 | Chromosome | 1,743,805 | 1,760,011 | 16,207 |
| Genomic_island_6 | Chromosome | 2,035,110 | 2,044,268 | 9159 |
| Genomic_island_7 | Chromosome | 2,256,066 | 2,260,915 | 4850 |
| Prophage_1 | Chromosome | 243,362 | 338,889 | 95,528 |
| Prophage_2 | Chromosome | 1,440,919 | 1,490,881 | 49,963 |
| Prophage_3 | Chromosome | 2,643,322 | 2,768,515 | 125,194 |
| Name | Location | Start | End | Repeat Number | Average Repeat Length (bp) | Spacer Number | Average Spacer Length (bp) |
|---|---|---|---|---|---|---|---|
| CRISPR.1 | Chromosome | 2,434,984 | 2,435,070 | 2 | 33 | 1 | 21 |
| CRISPR.2 | Chromosome | 2,604,343 | 2,604,427 | 2 | 19 | 1 | 47 |
| CRISPR.3 | Chromosome | 2,662,550 | 2,662,633 | 2 | 24 | 1 | 36 |
| CRISPR.4 | Plasmid | 51,317 | 51,420 | 2 | 28 | 1 | 48 |
| CRISPR.5 | Plasmid | 169,386 | 169,782 | 6 | 36 | 5 | 36 |
| Name | Gene Locus | Length (AA) | Annotation |
|---|---|---|---|
| Ferrous oxidation-related genes | |||
| Iro | AAE485_08910 | 90 | Iron oxidase, high potential iron–sulfur protein (HiPIP) family, secreted |
| RusB | AAE485_10410 | 189 | B-type rusticyanin, secreted, secreted |
| RusA | AAE485_10875 | 187 | A-type rusticyanin, secreted, secreted |
| Cyc1 | AAE485_10910 | 229 | c-type cytochrome, secreted, secreted |
| Cyc2 | AAE485_10915 | 487 | Outer membrane cytochrome c, secreted |
| CycA1 | AAE485_00725 | 260 | c-type cytochrome, secreted |
| Sulfur oxidation-related genes | |||
| SQR | AAE485_13635 | 394 | Sulfide:quinone reductase (SQR), FAD-dependent oxidoreductase, secreted |
| DoxA | AAE485_01905 | 172 | Thiosulfate:quinone oxidoreductase (TQO) subunit, DoxA domain-containing protein |
| DoxD | AAE485_08680 | 333 | Thiosulfate:quinone oxidoreductase (TQO) subunit, DoxD |
| CycA2 | AAE485_14115 | 237 | c-type cytochrome, secreted |
| TetH | AAE485_03710 | 503 | Tetrathionate hydrolase (TetH), PQQ-binding-like beta-propeller repeat protein, The Outer Membrane Protein Insertion Porin (Bam Complex) (OmpIP) Family, secreted |
| SoxB | AAE485_06770 | 575 | Thiosulfohydrolase SoxB, SignalP, secreted |
| SoxZ | AAE485_06780 | 110 | Thiosulfate oxidation carrier complex protein SoxZ, secreted |
| SoxY | AAE485_06785 | 170 | Thiosulfate oxidation carrier protein SoxY, secreted |
| Rho | AAE485_07140 | 141 | Rhodanese-like domain-containing protein, The Sulfate Permease (SulP) Family, secreted |
| Trx | AAE485_03190 | 194 | Thioredoxin_2 domain, secreted |
| Trx | AAE485_07965 | 206 | Disulfide isomerase, Thioredoxin_2 domain, secreted |
| Trx | AAE485_02165 | 394 | Pyridine nucleotide–disulfide oxidoreductase, secreted |
| Trx | AAE485_09980 | 406 | Pyridine nucleotide–disulfide oxidoreductase, secreted |
| Respiratory chain complex-related genes | |||
| CoxD | AAE485_10885 | 64 | Cytochrome C oxidase subunit IV, aa3 complex |
| CoxC | AAE485_10890 | 184 | cbb3-type cytochrome c oxidase subunit III, aa3 complex |
| CoxA | AAE485_10895 | 627 | cbb3-type cytochrome c oxidase subunit I, aa3 complex |
| CoxB | AAE485_10900 | 254 | Cytochrome C oxidase subunit II, aa3 complex |
| CoxD | AAE485_09335 | 184 | Cytochrome C oxidase subunit IV, aa3 complex |
| CoxC | AAE485_09330 | 627 | cbb3-type cytochrome c oxidase subunit III, aa3 complex |
| CoxA | AAE485_09325 | 254 | cbb3-type cytochrome c oxidase subunit I, aa3 complex |
| CoxB | AAE485_10900 | 254 | Cytochrome C oxidase subunit II, aa3 complex |
| CyoD | AAE485_12980 | 128 | Cytochrome O ubiquinol oxidase, bo3 complex |
| CyoC | AAE485_12985 | 212 | Cytochrome c oxidase subunit 3, bo3 complex |
| CyoB | AAE485_12990 | 715 | cbb3-type cytochrome c oxidase subunit I, bo3 complex |
| CyoA | AAE485_05135 | 310 | Ubiquinol oxidase subunit II, bo3 complex |
| CyoD | AAE485_13040 | 118 | Cytochrome O ubiquinol oxidase, bo3 complex |
| CyoC | AAE485_13045 | 211 | Cytochrome c oxidase subunit 3, bo3 complex |
| CyoB | AAE485_13050 | 677 | cbb3-type cytochrome c oxidase subunit I, bo3 complex |
| CyoA | AAE485_12995 | 321 | Ubiquinol oxidase subunit II, bo3 complex |
| Sdr1 | AAE485_00730 | 262 | SDR family NAD(P)-dependent oxidoreductase |
| PetA1 | AAE485_00735 | 206 | Ubiquinol–cytochrome c reductase iron–sulfur subunit, bc1 complex |
| PetB1 | AAE485_00740 | 402 | Cytochrome b N-terminal domain-containing protein subunit, bc1 complex |
| PetC1 | AAE485_00745 | 245 | Cytochrome c1 subunit, bc1 complex |
| Sdr2 | AAE485_03975 | 272 | SDR family NAD(P)-dependent oxidoreductase |
| PetA2 | AAE485_03980 | 206 | Ubiquinol–cytochrome c reductase iron–sulfur subunit, bc1 complex |
| PetB2 | AAE485_03985 | 404 | Cytochrome b N-terminal domain-containing protein subunit, bc1 complex |
| PetC2 | AAE485_03990 | 232 | Cytochrome c1 subunit, bc1 complex |
| Hip | AAE485_03995 | 104 | High potential iron–sulfur protein (HiPIP) family, secreted |
| Carbon fixation-related genes | |||
| rbcS | AAE485_12075 | 118 | Ribulose bisphosphate carboxylase (RubisCO) small subunit |
| rbcL | AAE485_04495 | 473 | Form I ribulose bisphosphate carboxylase (RubisCO) large subunit |
| rbcL | AAE485_09375 | 473 | Form I ribulose bisphosphate carboxylase (RubisCO) large subunit |
| rbcS | AAE485_04490 | 110 | Ribulose bisphosphate carboxylase (RubisCO) small subunit |
| rbcL | AAE485_12300 | 459 | Ribulose bisphosphate carboxylase (RubisCO) acetyl-CoA pathway |
| rbcS | AAE485_09380 | 110 | Ribulose bisphosphate carboxylase (RubisCO) small subunit |
| rbcL | AAE485_12080 | 473 | Form I ribulose bisphosphate carboxylase (RubisCO) large subunit |
| prkB | AAE485_06610 | 290 | Phosphoribulokinase |
| Nitrogen fixation-related genes | |||
| nifK | AAE485_09730 | 519 | Nitrogenase molybdenum–iron protein subunit beta |
| nifD | AAE485_09735 | 491 | Nitrogenase molybdenum–iron protein alpha chain |
| nifH | AAE485_09740 | 296 | Nitrogenase iron protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Liu, Y.; Liu, R.; Belqadi, W.; Zeng, W.; Yu, R.; Wu, X. Isolation, Sphalerite Bioleaching, and Whole Genome Sequencing of Acidithiobacillus ferriphilus QBS3 from Zinc-Rich Sulfide Mine Drainage. Life 2025, 15, 792. https://doi.org/10.3390/life15050792
Wang K, Liu Y, Liu R, Belqadi W, Zeng W, Yu R, Wu X. Isolation, Sphalerite Bioleaching, and Whole Genome Sequencing of Acidithiobacillus ferriphilus QBS3 from Zinc-Rich Sulfide Mine Drainage. Life. 2025; 15(5):792. https://doi.org/10.3390/life15050792
Chicago/Turabian StyleWang, Kan, Yuandong Liu, Run Liu, Wissal Belqadi, Weimin Zeng, Runlan Yu, and Xueling Wu. 2025. "Isolation, Sphalerite Bioleaching, and Whole Genome Sequencing of Acidithiobacillus ferriphilus QBS3 from Zinc-Rich Sulfide Mine Drainage" Life 15, no. 5: 792. https://doi.org/10.3390/life15050792
APA StyleWang, K., Liu, Y., Liu, R., Belqadi, W., Zeng, W., Yu, R., & Wu, X. (2025). Isolation, Sphalerite Bioleaching, and Whole Genome Sequencing of Acidithiobacillus ferriphilus QBS3 from Zinc-Rich Sulfide Mine Drainage. Life, 15(5), 792. https://doi.org/10.3390/life15050792







