Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals Key Trans-Target Genes Associated with Heat Stress Response in Rhododendron delavayi
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Heat Stress Treatment
2.2. Total RNA Extraction and Quality Examination
2.3. Library Construction for RNA-Seq
2.4. RNA-Seq Data Analysis
2.5. Analysis of DE-lncRNAs and DEGs
2.6. Function Annotation and Co-Expression Network Construction
2.7. qRT-PCR Analysis of Candidate DE-lncRNAs and DEGs
2.8. Statistical Analysis
3. Results
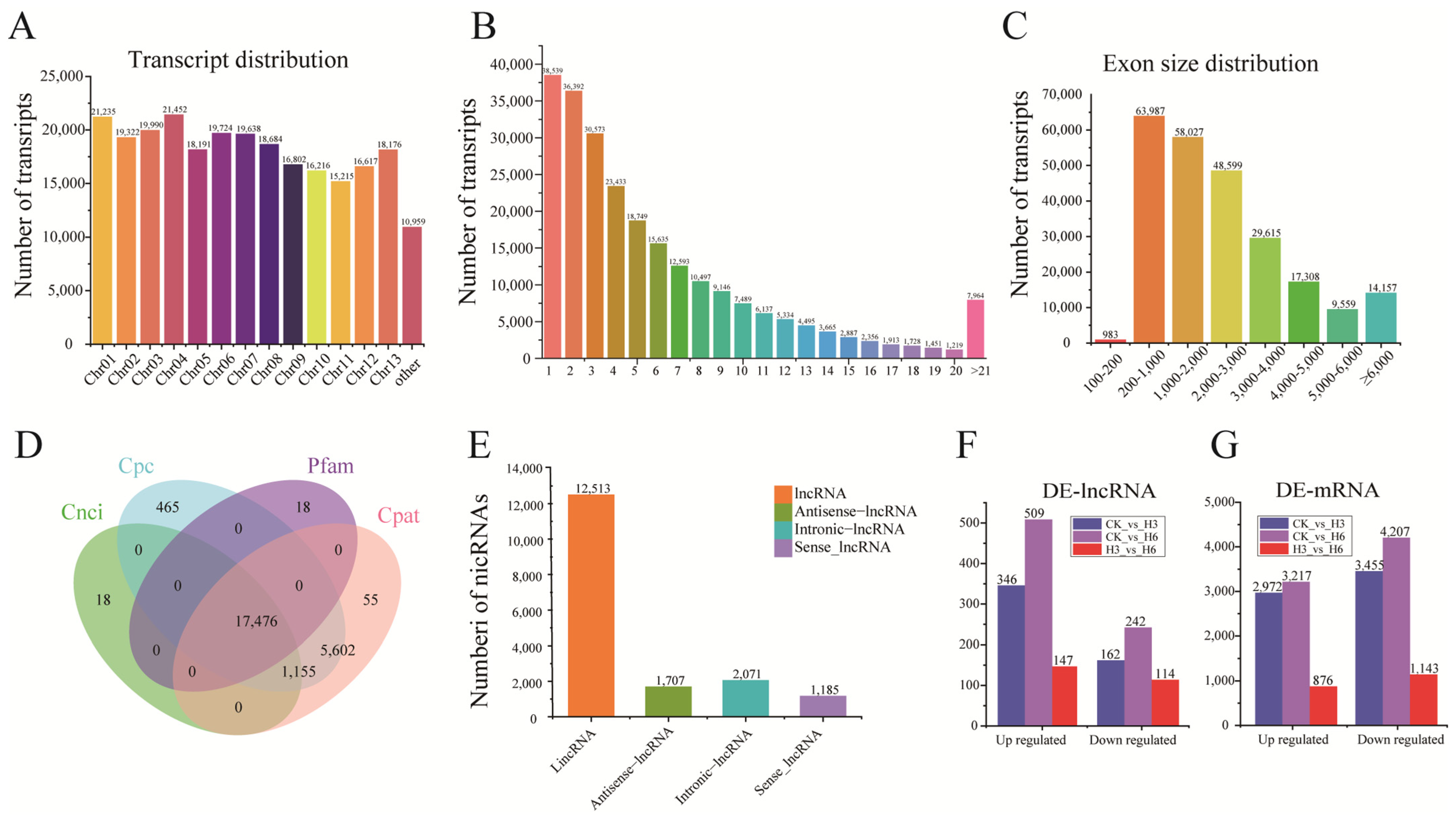
3.1. Comprehensive Overview of Transcriptome Sequencing (RNA-Seq)
3.2. Identification of lncRNAs and mRNA in R. delavayi Under Heat Stress Treatment
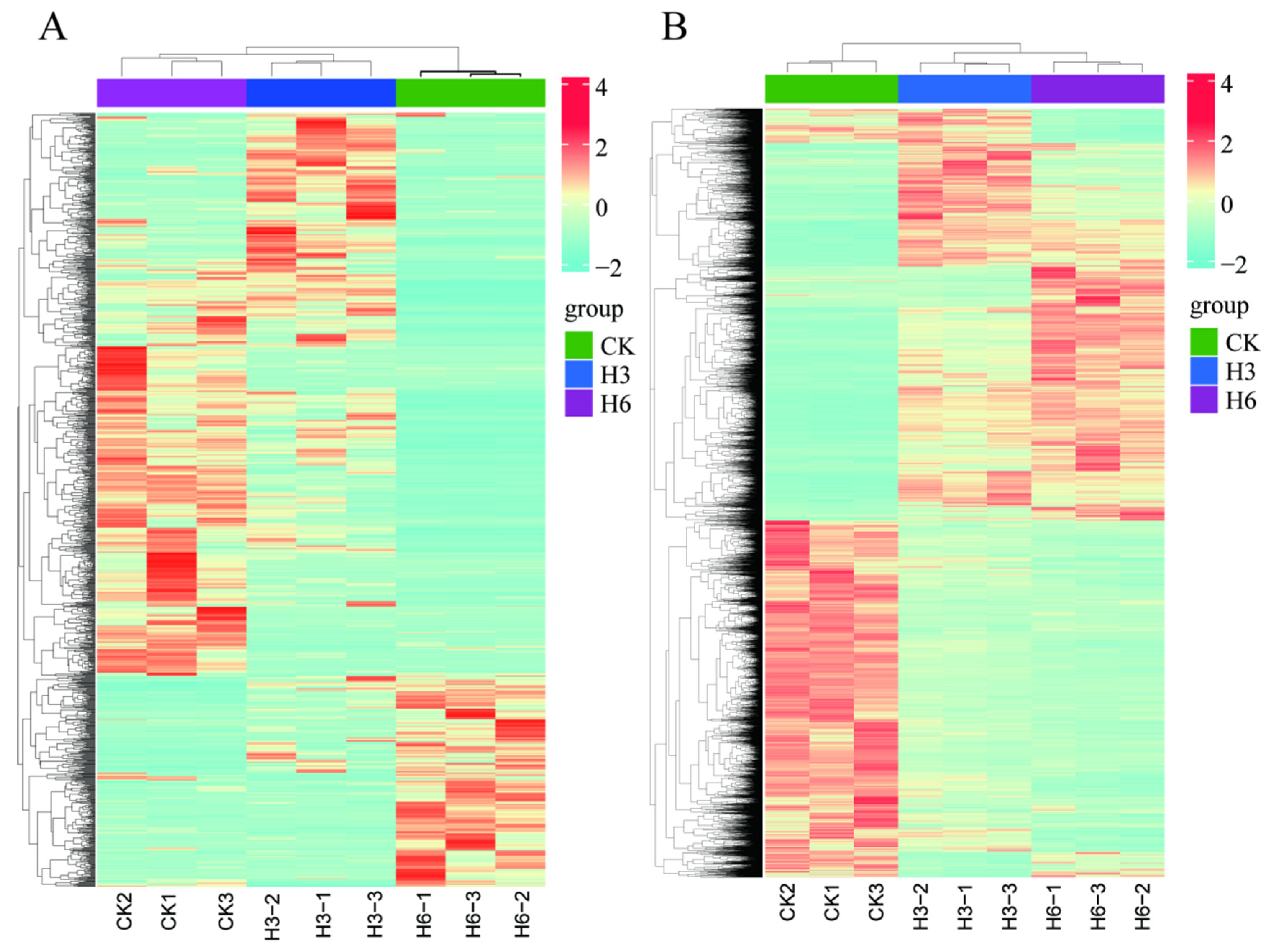
3.3. Identification of DE-lncRNAs and DEGs
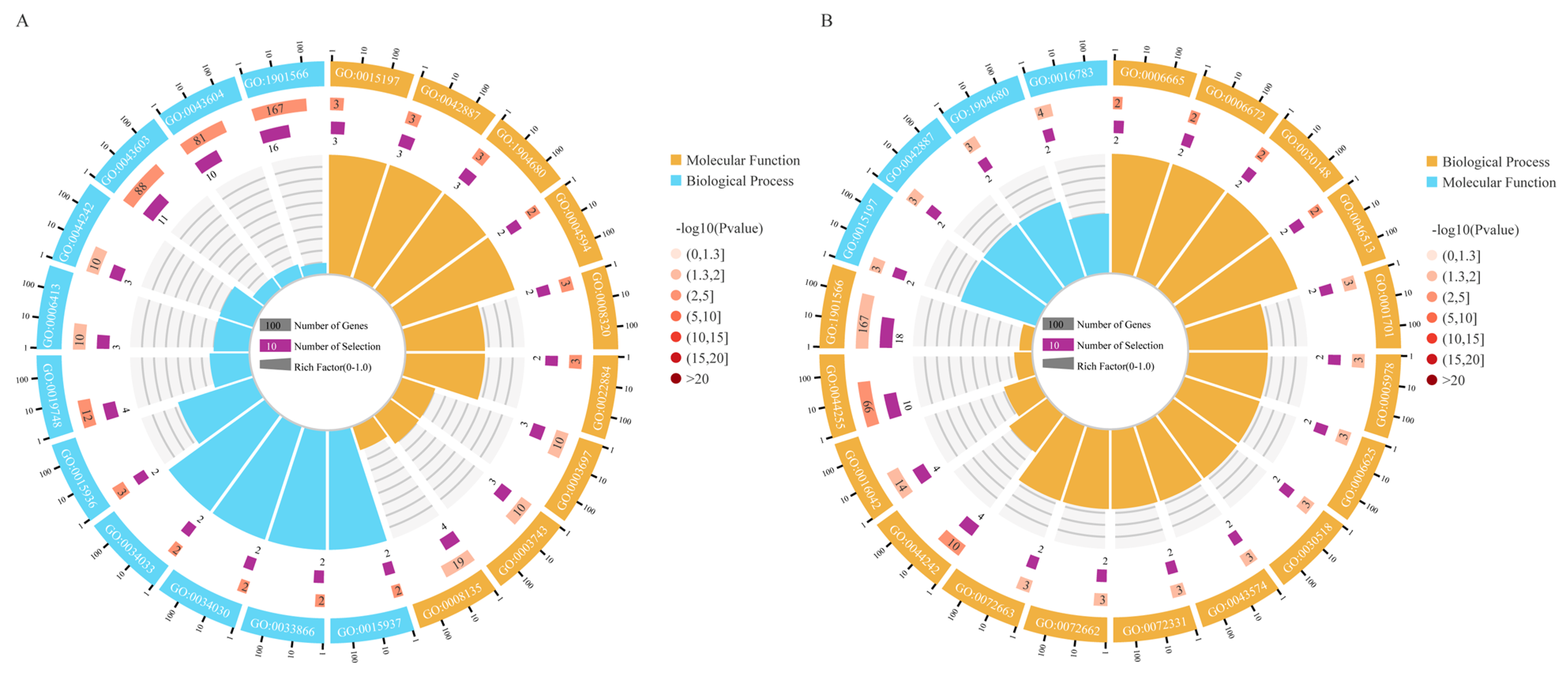
3.4. GO and KEGG Enrichment Analysis
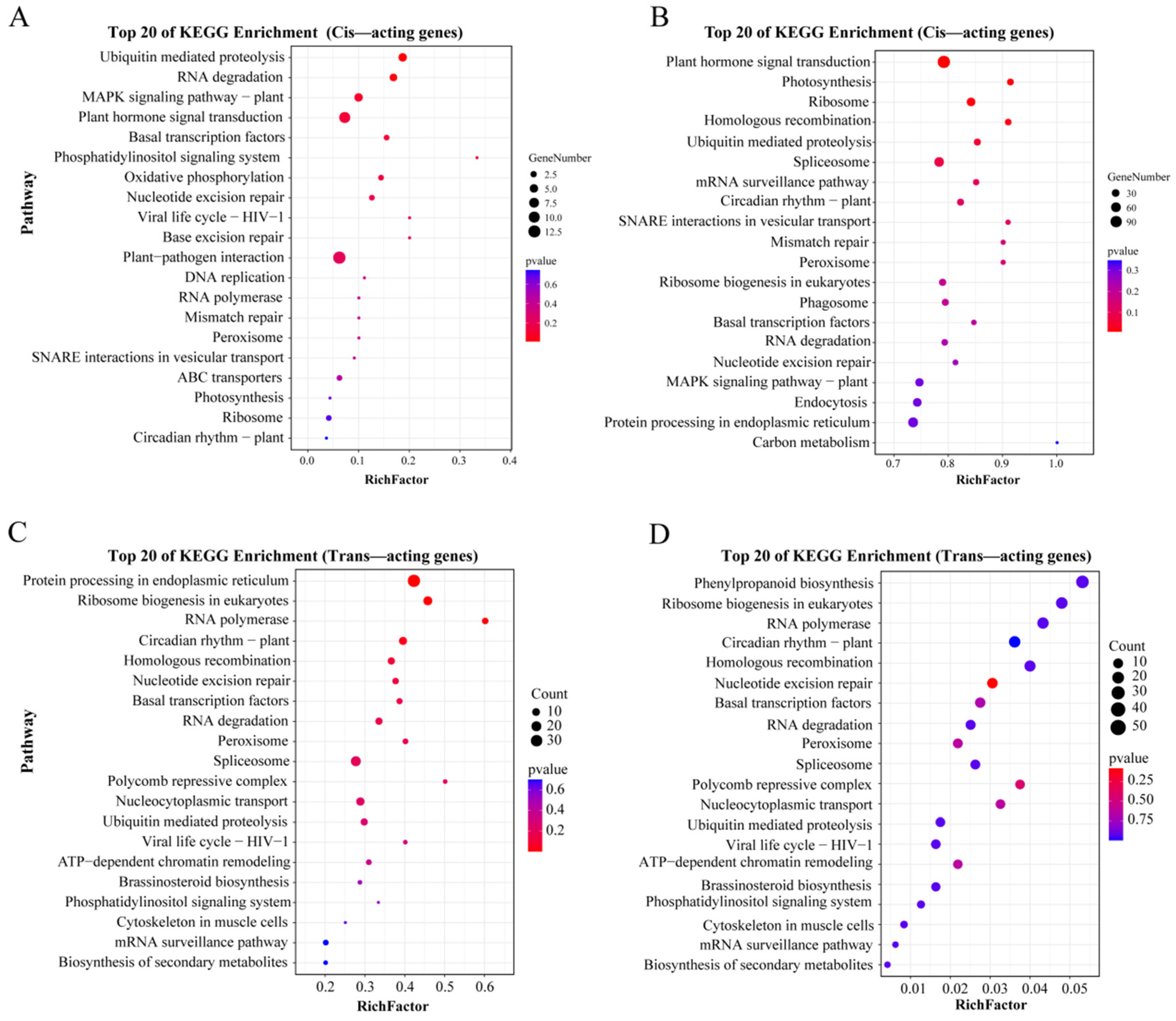
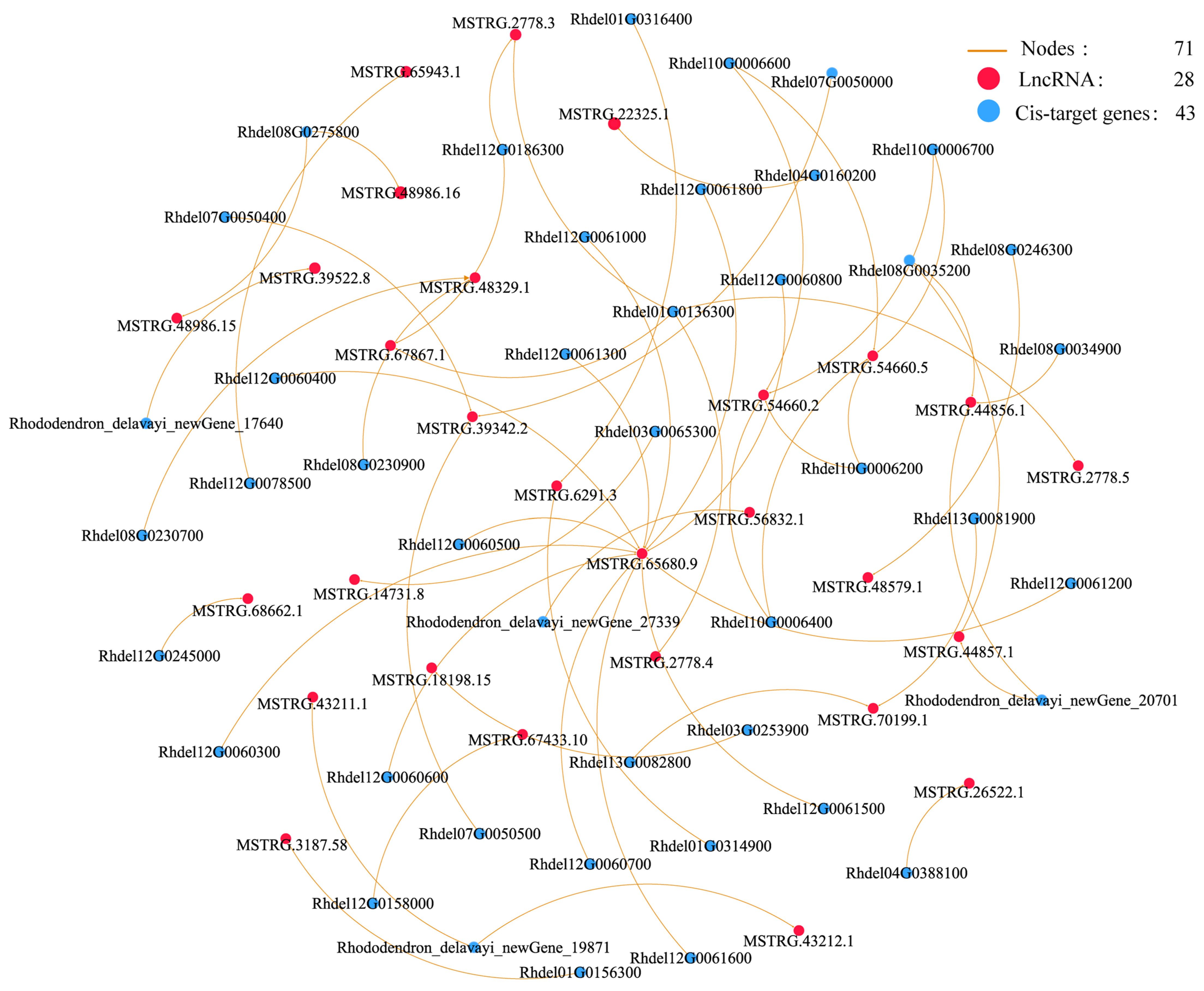
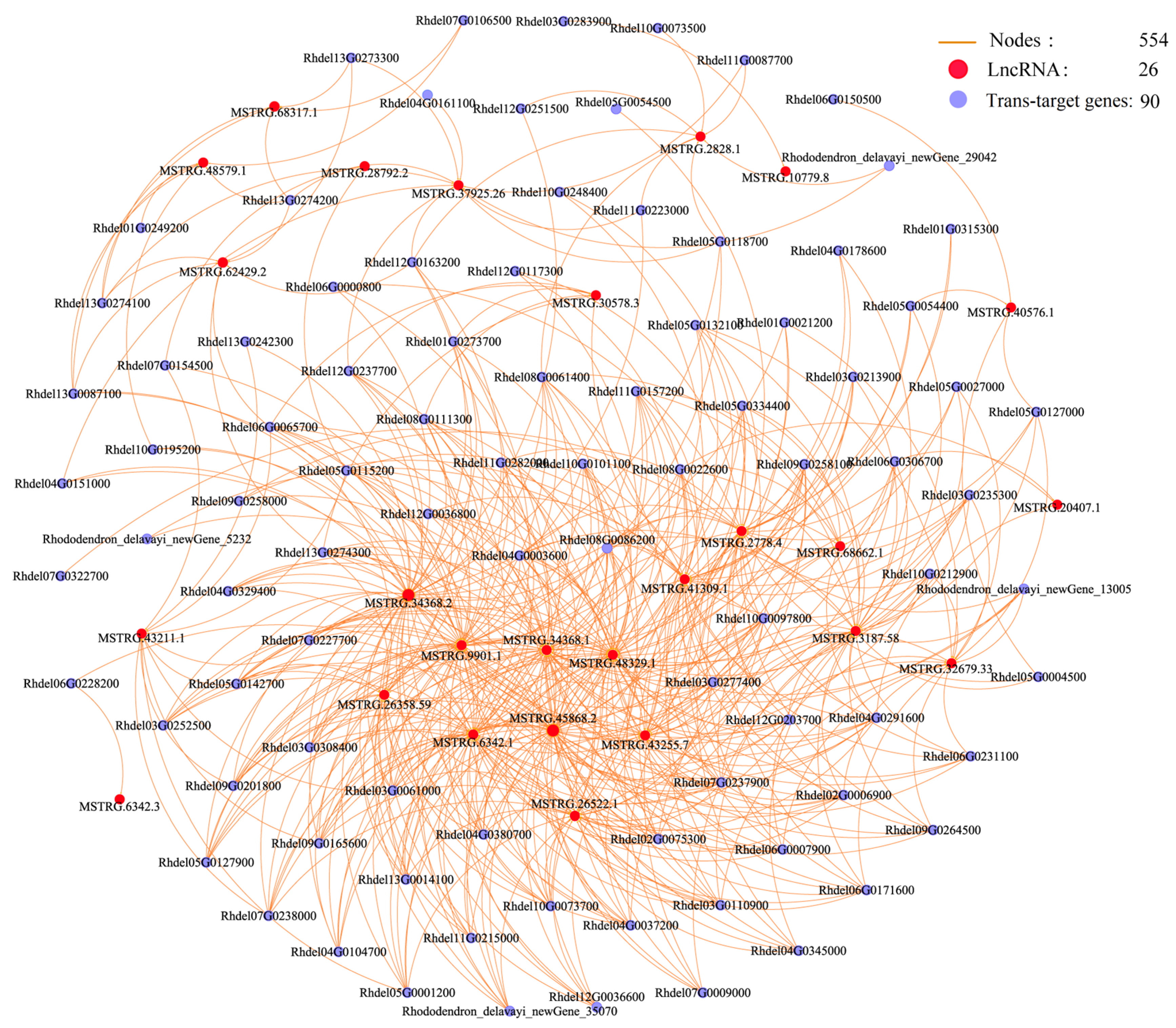
3.5. Co-Expression Networks of lncRNAs and Their Cis and Trans-Target Genes
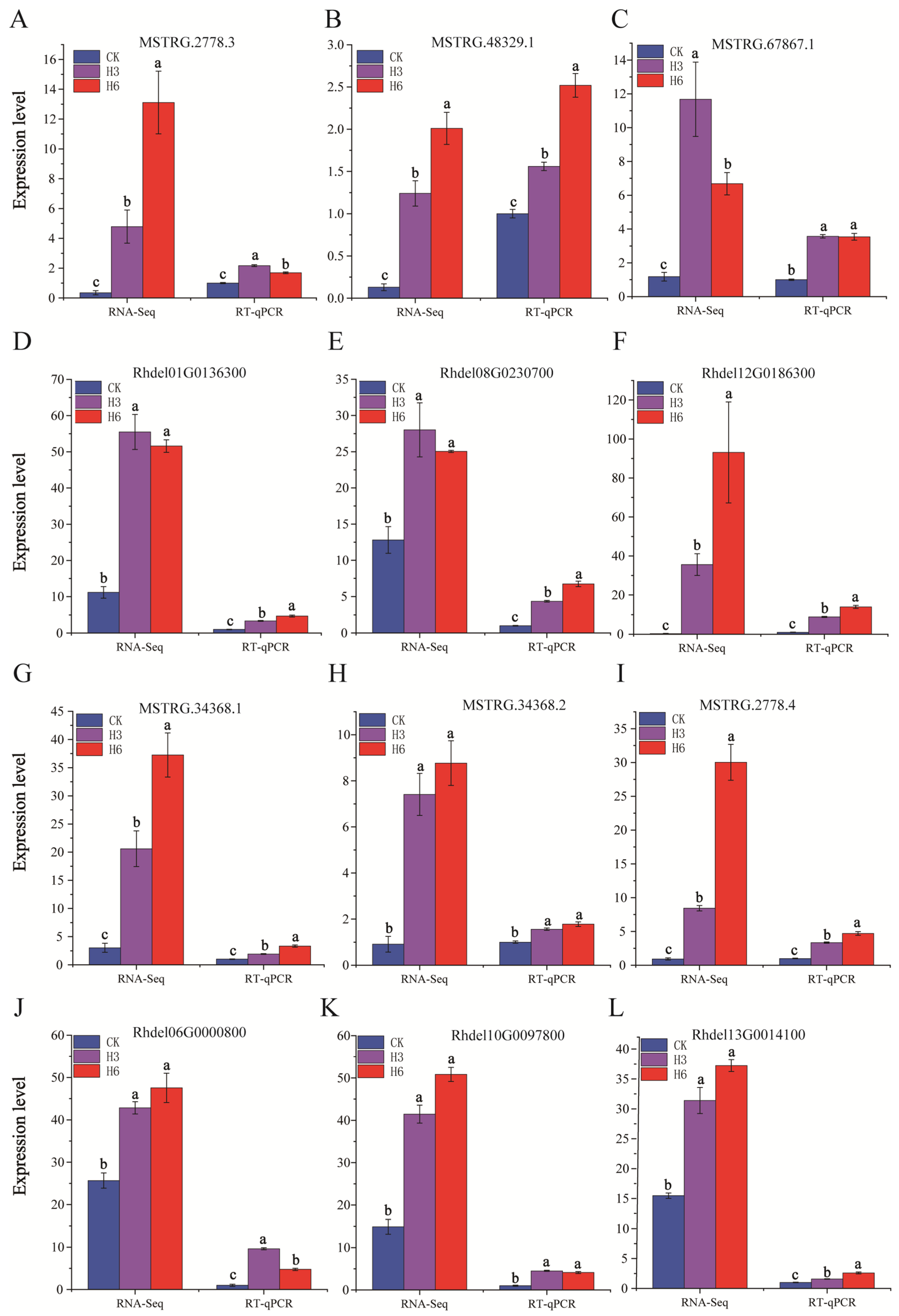
3.6. RT-qPCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Apostolova, E.L. Molecular Mechanisms of Plant Defense against Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 10339. [Google Scholar] [CrossRef] [PubMed]
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.; Ivanov, R.; Perrella, G.; Li, S. Multilevel Regulation of Abiotic Stress Responses in Plants. Front. Plant Sci. 2017, 8, 1564. [Google Scholar] [CrossRef]
- Manghwar, H.; Zaman, W. Plant Biotic and Abiotic Stresses. Life 2024, 14, 372. [Google Scholar] [CrossRef]
- Du, B.; Haensch, R.; Alfarraj, S.; Rennenberg, H. Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. Camb. Philos. Soc. 2024, 99, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.; Petrov, V. Reactive Oxygen Species and Abiotic Stress in Plants. Int. J. Mol. Sci. 2020, 21, 7433. [Google Scholar] [CrossRef] [PubMed]
- Balfagon, D.; Zandalinas, S.I.; Mittler, R.; Gomez-Cadenas, A. High temperatures modify plant responses to abiotic stress conditions. Physiol. Plant 2020, 170, 335–344. [Google Scholar] [CrossRef]
- Deng, K.; Li, Z.; Huang, T.; Huang, J. Noncoding RNAs in regulation of plant secondary metabolism. Plant Physiol. Biochem. 2024, 211, 108718. [Google Scholar] [CrossRef]
- Patra, G.K.; Gupta, D.; Rout, G.R.; Panda, S.K. Role of long non coding RNA in plants under abiotic and biotic stresses. Plant Physiol. Biochem. 2023, 194, 96–110. [Google Scholar] [CrossRef]
- Imaduwage, I.; Hewadikaram, M. Predicted roles of long non-coding RNAs in abiotic stress tolerance responses of plants. Mol. Hortic. 2024, 4, 20. [Google Scholar] [CrossRef]
- Yang, H.; Cui, Y.; Feng, Y.; Hu, Y.; Liu, L.; Duan, L. Long Non-Coding RNAs of Plants in Response to Abiotic Stresses and Their Regulating Roles in Promoting Environmental Adaption. Cells 2023, 12, 729. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Quan, M.; Liu, X.; Xiao, L.; Chen, P.; Song, F.; Lu, W.; Song, Y.; Zhang, D. Transcriptome analysis and association mapping reveal the genetic regulatory network response to cadmium stress in Populus tomentosa. J. Exp. Bot. 2021, 72, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhao, H.; Niu, Y.; Wang, Y. Long noncoding RNA from Betula platyphylla, BplncSIR1, confers salt tolerance by regulating BpNAC2 to mediate reactive oxygen species scavenging and stomatal movement. Plant Biotechnol. J. 2024, 22, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zheng, H.; Li, J.; Liu, L.; Zhang, X.; Sui, N. Comparative Transcriptome Analysis Reveals New lncRNAs Responding to Salt Stress in Sweet Sorghum. Front. Bioeng. Biotechnol. 2020, 8, 331. [Google Scholar] [CrossRef]
- Wang, A.; Hu, J.; Gao, C.; Chen, G.; Wang, B.; Lin, C.; Song, L.; Ding, Y.; Zhou, G. Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp. chinensis). Sci. Rep. 2019, 9, 5002. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, Y.; Pu, S.; Zhang, X.; Li, Z.; Chen, J. Third-generation sequencing found LncRNA associated with heat shock protein response to heat stress in Populus qiongdaoensis seedlings. BMC Genom. 2020, 21, 572. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Li, W.; Wang, S.; Hao, L.; Xu, C.; Yu, Y.; Xiang, L.; Li, T.; Jiang, F. A long noncoding RNA HILinc1 enhances pear thermotolerance by stabilizing PbHILT1 transcripts through complementary base pairing. Commun. Biol. 2022, 5, 1134. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Y.; Wu, X.; Wen, X.; Li, D.; Zhou, H.; Li, Z.; Liu, B.; Wei, J.; Chen, F.; et al. High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol. J. 2021, 19, 2544–2560. [Google Scholar] [CrossRef]
- Wang, X.-J.; Wei, Y.-F.; Liu, Z.; Yu, T.; Fu, Y.-H.; Song, X.-M. TEGR: A comprehensive Ericaceae Genome Resource database. J. Integr. Agric. 2023, 24, 1140–1151. [Google Scholar] [CrossRef]
- Liu, S.J.; Cai, C.; Cai, H.Y.; Bai, Y.Q.; Wang, D.Y.; Zhang, H.; Peng, J.G.; Xie, L.J. Integrated analysis of transcriptome and small RNAome reveals regulatory network of rapid and long-term response to heat stress in Rhododendron moulmainense. Planta 2024, 259, 104. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Peters, M.; Mueller, R.O. Correlational analysis of ordinal data: From Pearson’s r to Bayesian polychoric correlation. Asia Pac. Educ. Rev. 2010, 11, 459–466. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, 900. [Google Scholar] [CrossRef]
- Baev, V.; Gisel, A.; Minkov, I. The Fascinating World of Plant Non-Coding RNAs. Int. J. Mol. Sci. 2023, 24, 10341. [Google Scholar] [CrossRef]
- Song, L.; Fang, Y.; Chen, L.; Wang, J.; Chen, X. Role of non-coding RNAs in plant immunity. Plant Commun. 2021, 2, 100180. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-X.; Zhang, J.; Wan, Z.-Y.; Huang, S.-X.; Di, H.-C.; He, Y.; Jin, S.-H. Physiological and transcriptome analyses provide new insights into the mechanism mediating the enhanced tolerance of melatonin-treated rhododendron plants to heat stress. J. Integr. Agric. 2023, 22, 2397–2411. [Google Scholar] [CrossRef]
- He, X.; Guo, S.; Wang, Y.; Wang, L.; Shu, S.; Sun, J. Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.). Physiol. Plant 2020, 168, 736–754. [Google Scholar] [CrossRef]
- Bhatia, G.; Singh, A.; Verma, D.; Sharma, S.; Singh, K. Genome-wide investigation of regulatory roles of lncRNAs in response to heat and drought stress in Brassica juncea (Indian mustard). Environ. Exp. Bot. 2020, 171, 103922. [Google Scholar] [CrossRef]
- Arunkumar, G. LncRNAs: The good, the bad, and the unknown. Biochem. Cell Biol. 2024, 102, 9–27. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, W.; Wang, M.; Zhou, X. The Role of Long Noncoding RNAs in Gene Expression Regulation. Gene Expr. Profiling Cancer 2019, 5, 17. [Google Scholar] [CrossRef]
- Sun, H.F.; Wang, X.N.; Li, Y.N.; Wang, L.L.; Li, Y.Y.; Ma, L.J.; Li, X.M. Long non-coding RNAs modulate glutathione metabolism gene expression and tolerance to Pb stress in root tissue of endophyte-infected rice seedling. Ecotoxicol. Environ. Saf. 2025, 291, 117872. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, S.; Fu, H.; Zhou, C.; Chen, L.; Li, X.; Lin, Y.; Lai, Z.; Guo, Y. Transcriptome and Phytochemical Analyses Provide New Insights Into Long Non-Coding RNAs Modulating Characteristic Secondary Metabolites of Oolong Tea (Camellia sinensis) in Solar-Withering. Front. Plant Sci. 2019, 10, 1638. [Google Scholar] [CrossRef]
- Dong, C.; Peng, X.; Yang, X.; Wang, C.; Yuan, L.; Chen, G.; Tang, X.; Wang, W.; Wu, J.; Zhu, S.; et al. Physiological and Transcriptomic Responses of Bok Choy to Heat Stress. Plants 2024, 13, 1093. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, L.; Rusalepp, L.; Kaurilind, E.; Sulaiman, H.Y.; Pussa, T.; Niinemets, U. Heat priming improved heat tolerance of photosynthesis, enhanced terpenoid and benzenoid emission and phenolics accumulation in Achillea millefolium. Plant Cell Environ. 2021, 44, 2365–2385. [Google Scholar] [CrossRef]
- Wang, H.; Xu, W.; Zhang, X.; Wang, L.; Jia, S.; Zhao, S.; Li, W.; Lu, R.; Ren, A.; Zhang, S. Transcriptomics and metabolomics analyses of Rosa hybrida to identify heat stress response genes and metabolite pathways. BMC Plant Biol. 2024, 24, 874. [Google Scholar] [CrossRef]
- Wu, X.; Ma, Y.; Wang, P.; Wu, J.; Li, N.; Zhang, Z.; Xie, R.; Wang, D.; Nie, H. Transcriptome sequencing and screening of anthocyanin related genes in purple potato tubers (Solanum tuberosum L.). BMC Genom. 2024, 25, 1159. [Google Scholar] [CrossRef]
- Abiko, M.; Akibayashi, K.; Sakata, T.; Kimura, M.; Kihara, M.; Itoh, K.; Asamizu, E.; Sato, S.; Takahashi, H.; Higashitani, A. High-temperature induction of male sterility during barley (Hordeum vulgare L.) anther development is mediated by transcriptional inhibition. Sex. Plant Reprod. 2005, 18, 91–100. [Google Scholar] [CrossRef]
- Dard, A.; Weiss, A.; Bariat, L.; Auverlot, J.; Fontaine, V.; Picault, N.; Pontvianne, F.; Riondet, C.; Reichheld, J.P. Glutathione-mediated thermomorphogenesis and heat stress responses in Arabidopsis thaliana. J. Exp. Bot. 2023, 74, 2707–2725. [Google Scholar] [CrossRef]
- Hewitt, S.; Hernandez-Montes, E.; Dhingra, A.; Keller, M. Impact of heat stress, water stress, and their combined effects on the metabolism and transcriptome of grape berries. Sci. Rep. 2023, 13, 9907. [Google Scholar] [CrossRef]
- Zheng, L.; Wu, W.; Chen, Q.; Zhang, G.; Gao, F.; Zhou, Y. Integrated transcriptomics, proteomics, and metabolomics identified biological processes and metabolic pathways involved in heat stress response in jojoba. Ind. Crops Prod. 2022, 183, 114946. [Google Scholar] [CrossRef]
- Zhao, X.; Li, P.; Liu, X.; Xu, T.; Zhang, Y.; Meng, H.; Xia, T. High temperature increased lignin contents of poplar (Populus spp.) stem via inducing the synthesis caffeate and coniferaldehyde. Front. Genet. 2022, 13, 1007513. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.T.; Zhang, L.L.; Han, J.J.; Zhou, M.; Liu, J.X. Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat-stress gene expression during recovery in Arabidopsis. Plant J. 2021, 105, 1326–1338. [Google Scholar] [CrossRef]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef]
| Sample | Clean Reads | Mapped Reads | Uniq Mapped Reads | Multiple Mapped Reads | GC (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| CK1 | 72,796,176 | 65,816,206 (90.41%) | 59,540,530 (81.79%) | 6,275,676 (8.62%) | 44.11 | 94.4 |
| CK2 | 66,000,982 | 60,399,258 (91.51%) | 54,745,987 (82.95%) | 5,653,271 (8.57%) | 44.45 | 94.69 |
| CK3 | 72,965,128 | 67,690,520 (92.77%) | 62,163,128 (85.20%) | 5,527,392 (7.58%) | 44.39 | 95.1 |
| H3-1 | 61,128,826 | 54,812,099 (89.67%) | 50,337,543 (82.35%) | 4,474,556 (7.32%) | 44.68 | 94.28 |
| H3-2 | 63,427,652 | 56,640,331 (89.30%) | 52,148,500 (82.22%) | 4,491,831 (7.08%) | 44.53 | 94.55 |
| H3-3 | 70,936,282 | 63,721,989 (89.83%) | 58,902,339 (83.04%) | 4,819,650 (6.79%) | 44.44 | 94.3 |
| H6-1 | 60,341,510 | 54,375,726 (90.11%) | 50,600,561 (83.86%) | 3,775,165 (6.26%) | 44.27 | 94.28 |
| H6-2 | 75,965,538 | 68,870,447 (90.66%) | 63,723,386 (83.88%) | 5,147,061 (6.78%) | 44.34 | 94.24 |
| H6-3 | 69,575,994 | 62,557,432 (89.91%) | 57,860,201 (83.16%) | 4,697,231 (6.75%) | 44.9 | 94.62 |
| LncRNA | Target Genes | KEGG Pathways | Gene Annotation | Abbr. |
|---|---|---|---|---|
| MSTRG.2778.3 | Rhdel01G0136300 | RNA polymerase | RNA polymerase subunit | PPABC1 |
| MSTRG.48329.1 | Rhdel08G0230700 | Phenylpropanoid synthesis | Cinnamoyl-CoA reductase 1 | CCR |
| MSTRG.67867.1 | Rhdel12G0186300 | Phenylpropanoid synthesis | UDP-glucosyltransferas | UGT89B2 |
| MSTRG.34368.2 | Rhde106G0000800 | Val, leu, Ile degradation | Trithorax-related proteins | TrxG |
| MSTRG.2778.4 | Rhde110G0097800 | Linoleic acid metabolism | Phosphoenolpyruvate carboxylase | PEPC |
| MSTRG.34368.1 | Rhde113G0014100 | Val, leu, Ile degradation | Isocitrate dehydrogenase | IDH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, Y.; Zhang, X.; An, L.; Tian, Z. Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals Key Trans-Target Genes Associated with Heat Stress Response in Rhododendron delavayi. Life 2025, 15, 697. https://doi.org/10.3390/life15050697
Liu C, Liu Y, Zhang X, An L, Tian Z. Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals Key Trans-Target Genes Associated with Heat Stress Response in Rhododendron delavayi. Life. 2025; 15(5):697. https://doi.org/10.3390/life15050697
Chicago/Turabian StyleLiu, Changming, Yang Liu, Xinyue Zhang, Lujie An, and Zhiguo Tian. 2025. "Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals Key Trans-Target Genes Associated with Heat Stress Response in Rhododendron delavayi" Life 15, no. 5: 697. https://doi.org/10.3390/life15050697
APA StyleLiu, C., Liu, Y., Zhang, X., An, L., & Tian, Z. (2025). Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals Key Trans-Target Genes Associated with Heat Stress Response in Rhododendron delavayi. Life, 15(5), 697. https://doi.org/10.3390/life15050697





